This post is a great example of how to get the most from your Y-DNA. In this case we will not only prove a Y-DNA relationship, but also pinpoint the exact location in Ireland where a man's ancestors lived.
I will explain the different types of Y-DNA tests, how to do your own evaluations of the results, and give a list of further suggestions on how to get the most from your results and what to do next. This will be a long post.
Can we find immigrant origins when there may be no records?
My father is very proud of his Irish heritage, but he wants to know more. From family tradition he knows that his ancestor, William Gibbs, emigrated to Canada from Ireland.
That's the only information that he knew about his Irish origins. Can we find out more?
What we know from Canadian records
William Gibbs is first listed in the 1851 census of Sheffield, Addington County, Ontario. His wife was Mary Coulter, and he is listed in the census next to her parents.
The census states that William, his wife, and daughter Mary were all born in Ireland.
 |
| Gibbs family in 1841 census of Sheffield, Ontario, Canada |
William is listed in the 1861 census of Sheffield, but this time the census states that all of his children, including his first daughter, were born in Canada.
 |
| Gibbs family in the 1861 census of Sheffield, Ontario, Canada |
Other censuses, and the death record of the daughter Mary, confirm that all children were born in Ontario.
It appears that William may have come to Canada around the time of The Great Famine. There is no passenger list to tell us exactly when he arrived.
No record has been found that contains a date of birth or the names of his parents.
Luckily, however, his death record [Number 18 in the image below] states that he was born in County Monaghan, Ireland.
 |
| Death of William Gibbs 15 Jun 1882 in Sheffield, Ontario, Canada |
So here's all we know from Canadian records about William's origins: William Gibbs was born around 1826 in County Monaghan, Ireland, and was in Canada by 1851.
Searching records in Ireland
First, as you may know, there is a scarcity of Irish records. In many cases we are stuck with just knowing that our ancestor came from Ireland. But that didn't deter me!
After seeing that William was born in County Monaghan I immediately began searching through the records that remain.
One of the most useful records for the time when William would have been living in Ireland are the Tithe Applotments. These records were taken between 1823-1837 and can be used as a census substitute.
In the entire County of Monaghan, there was not a single Gibbs family. I could not find the family in other surviving records, so I eventually gave up.
An unexpected message
I came to stay with my parents in 2019 to help take care of my mother. She died Christmas morning. I was helping my father when the Coronavirus quarantine was announced, so then I stayed.
In March of this year he was talking about his family and said, "About 10 years ago, a man named Jebb contacted me from Ireland and said he thought we might be related." I asked, "What made him think that?" He replied, "DNA."
I asked if he still had the email, and he said he didn't, but he thought that he could find the man's email address. A few minutes later I was sending an email to a man in Ireland using an old email address and hoping that he still used it.
The next day Mr. Jebb replied. He lives in Ireland, and his ancestors were from the townland of Billis, County Monaghan, Ireland.
Jebb DNA
Mr. Jebb informed me that many years ago he had done a Y-DNA test with Ancestry.com [Ancestry no longer conducts Y-DNA tests].
Other Jebb men had tested at the same company, and some of them were matches. He said he was also a distant match to Gibbs.
He was the only Jebb had also taken a 25-marker Y-DNA test with Family Tree DNA, but he did not have any Jebb or Gibbs matches in the FTDNA database.
At the time these men were tested, most companies had only one kind of test available for Y-DNA. This was the STR test.
What is an STR test?
"STR" stands for Short Tandem Repeat. A Short Tandem Repeat means that a short series of bases (nucleotides) is repeated side-by-side several times, something like a hiccup.
Here's an STR example: AGATAGATAGATAGATAGATAGATAGATAGAT. In this example, the sequence "AGAT" is repeated eight times.
There are many known locations on the Y-chromosome where these Short Tandem Repeats occur. Each of these locations is identified by a marker.
Each marker is given a name such as DYS393. This abbreviation stands for DNA Y-chromosome Segment 393. At this marker the number of repeats is counted and reported.
So, for example, in a 12-marker test the testing company will examine 12 named locations on the Y-chromosome and report the number of repeats at each location.
Old Y-DNA tests
After hearing that Jebb had tested at Ancestry and at Family Tree DNA (FTDNA) I was in quite a quandary. My father had not taken Y-DNA tests with either Ancestry.com or with FTDNA.
However, he had taken two Y-DNA tests. In 2006 my father had taken a 12-marker Y-DNA test from National Geographic's Genographic Project. I had transferred the results to Family Tree DNA.
He had also taken a 43-marker Y-DNA test from the Sorenson Molecular Genealogy Foundation (SMGF). Although SMGF did not display the results, they did publish their database online. As a person guessed at each marker correctly it would show up in the results. I did exactly that, until all results were showing, and then transferred the SMGF results to Ancestry.com and to YSearch.org.
SMGF tested 43 markers, but one of these markers was not tested by Family Tree DNA, so only 42 markers are recorded in the YSearch record.
You would think I'd be safe with the DNA in that many databases. But, unfortunately, all but one of them is now gone.
Ancestry.com purchased SMGF in 2010, and removed the SMGF online database. Ancestry then discontinued Y-DNA testing and removed its own Y-DNA database.
Family Tree DNA removed the YSearch database. This one was the most devastating because I had tested the Y-DNA of hundreds of men. The Y-DNA test of every man from any company had been entered into YSearch, and all were now gone.
Most recently, the Genographic Project has been discontinued, and its database has been taken down.
So what did I have left? I had the 12-marker Genographic Project results that had been transferred to Family Tree DNA, and one more thing--I had taken a screenshot of the Gibbs YSearch results.
 |
| Gibbs 42-marker STR results recorded at YSearch.org |
I could compare the Jebb Y-DNA results at Family Tree DNA with the Gibbs 12-marker results that had been transferred there.
I immediately looked to see if Jebb was on the Gibbs list of 12-marker Y-DNA matches at Family Tree DNA. He wasn't. Right off the bat, Family Tree DNA is indicating that these two men are not related.
If they aren't showing up as matches at only 12 markers, this is not a good sign. But, I still had my YSearch screenshot, and Mr. Jebb allowed me access to his 25-marker Y-DNA results:
 |
| Jebb 25-marker STR results |
By comparing the two we can see how many differences there are.
There is already a difference at the second marker, DYS390, where Gibbs has 24, and Jebb has 23.
The second difference is at DYS385b where Gibbs has 16, and Jebb has 15. So these two men have two differences in the first 12 markers.
A third difference is at DYS447 where Gibbs has 26 and Jebb has 25. This is definitely not looking promising.But Mr. Jebb also informed me that he had compiled a database of his Ancestry Y-DNA results comparing them to the results of any possible matches. He sent me the list, and I carefully compared them.
In addition to his own results there were three matches named Jebb on his list, and my father's Y-DNA results matched all of the men more closely than his results matched Mr. Jebb's. For example, all three of the Jebb matches had a 24 at DYS390. So there was a chance!
I wrote back to Mr. Jebb and told him that our best shot of solving this was to upgrade both the Jebb and Gibbs Y-DNA tests. I said that it was possible that no matter how many STRs we ordered, these two men might still not show up as matches, but if they were really related a Big Y test would prove it.
The most recent version of the Big Y test is called Big Y-700. This test will prove the Jebb-Gibbs relationship because not only does it include more than 700 STRs, it includes the very important SNPs. In addition, the Big Y-700 includes a separate test of the first 111 basic STRs.
What is a SNP?
SNP (pronounced "snip") stands for Single Nucleotide Polymorphism. It occurs when a single base (nucleotide) mutates.
In the image below the ancestral nucleotide A has mutated to a T in Man 1. [See the fifth letter from the left.]
 |
| SNPs and STRs (Image by Mark Jobling) |
The main difference between SNPs and STRs is their stability.
STRs can mutate back and forth. For example, in the image above we don't know which of the repeat values came first.
Man 1 has CTA repeated 5 times, but CTA is repeated 6 times in Man 2's results and 7 times in Man 3's results.
Which is the ancestral value--was it first a 5 that changed to a 6 in Man 2 and then a 7 in Man 3? Or was it first a 6 that mutated down to a 5 in Man 1 and up to a 7 in Man 3? Or was it first a 7 that mutated down in Men 1 and 2? There is no way to tell.
Furthermore, these not the only three options--STRs can mutate up in one generation and back down in another generation making two men look more closely related than they really are.
SNPs, on the other hand, are generally one-time events. When a SNP occurs it is passed down to all future generations.
Some SNPs are proven to have occurred thousands of years ago. Others occurred in recent times.
When a new SNP is found through Y-DNA testing it is given a name and placed on the human Y-DNA tree (called the haplotree). The name of this SNP can become the name of a new haplogroup.
A haplogroup is simply a group of men who share a SNP. SNP testing is progressing so rapidly that SNPs occurring in the genealogical time period are being now placed onto the Y-DNA haplotree.
The Big Y test is ordered
The day after I suggested upgrading to Mr. Jebb, I received a reply stating that he had ordered the Big Y on March 7.
I normally wait until the Big Y is on sale to upgrade, but I thought that if I ordered the test right at that time, the 111 STRs might be completed by Father's Day. That would be the best Father's Day present ever if I could show my dad that he was very likely related to Mr. Jebb.
So I immediately ordered the Big Y test.
Back to the records
I spent the next few months working almost non-stop reading any records relating to the Jebb family. In case the Y-DNA results showed that these men were really related, I wanted to find the possible Gibbs ancestor (which seemed impossible).
I read every Jebb deed, went through surviving church records, etc.
For example, although there were no Gibbs families in the Tithe Applotments for County Monaghan there were Jebb families there.
The Tithe Applotments can be searched at The National Archives of Ireland where we see that there were fourteen Jebb families listed:
 |
| Index to Tithe Applotments at The National Archives of Ireland |
I wanted to trace all of the Jebb emigrants to see which ones ended up in Canada. I also wanted to find descendants of multiple Jebb lines for possible future Y-DNA tests.
This was much more work than I had ever imagined because multiple Jebb descendants had left County Monaghan and emigrated to Scotland, England, India, Australia, Canada, and the United States. Many of them had migrated through multiple countries. I was going through the records in all of these countries compiling lists of Jebb families.
I eliminated many Jebb men as the possible ancestors of William Gibbs, and narrowed it down to just a few. One of them is the best candidate, but since there is no baptismal or other record for William Gibbs the proof can only be provided with Y-DNA.
Y-STR Results
The 111 STR results came in on June 10. Father's Day was June 21, so I had a few days to prepare the presentation of records and to show my father the most exciting news--the DNA results.
At 111 markers Mr. Jebb was my father's only match:
 |
| STR match list |
In the Genetic Distance Column you see the number 5. This means that there were five differences between the two men in the 111 markers. We know that two of them occurred within the first 12 markers, and that would normally indicate that these two men are not related.
The 111 STRs indicate that these men probably are related, and if so, the SNPs will prove it.
Big Y results
Mr. Jebb's Big Y results came in first. I checked every day to see my father's results, but they did not show. I called Family Tree DNA, and they said that his test had failed and had to be redone. Mr. Jebb had no matches yet.
The Big Y testing is done in two stages, the first part is automated, and the second stage involves a manual review of the results to more accurately identify the SNPs, name them, and place them on the haplotree.
At the end of the automated process, you will see results that may be later modified by the manual review. Family Tree DNA will place each person on a tree called the Block Tree which is a tree showing where that man belongs in the tree of mankind from the earliest ancestors down to the most recent ones.
After logging into the Family Tree DNA account, there is a Big Y section for those who have taken the test. Click "Block Tree" to see your placement on the tree.
 |
| Big Y menu: Click Block Tree |
When you first click on the Block tree, you will see this welcome message with an explanation of the tree:
 |
| Block Tree Welcome message |
Click "Show me around" to see how to navigate the tree, then see the tree itself.
For people who have not taken the Big Y test, the public Block Tree can be accessed by going to the Family Tree DNA website.
Then go to the bottom of the page and click "Y-DNA Haplotree." You can then search for any position on the tree.
 |
| Family Tree DNA Y-DNA Haplotree |
Jebb's position on the Block Tree
Mr. Jebb appeared on the Big Y Block Tree in a branch called R-BY50723:
 |
| Branch R-BY50723 in Block Tree |
At the top of the above screenshot is a white block [your own branch of the Block Tree is always shown in white] containing 26 SNPs with names like BY50723, BY137843, etc. They are grouped together because the order in which these occurred is not yet known.
When a new Big Y tester has some of these SNPs, but not others, the ones that are shared by the new tester will then be known to have occurred in earlier generations than the SNPs that are not shared.
As more people test, it is possible for many of these SNPs to be placed in generational order on the tree.
The SNPs beginning with the letters BY were discovered with earlier Big Y tests, and the ones beginning with the letters FT were discovered with the more recent Big Y-700 test (which is the test Jebb and Gibbs took).
I looked at Mr. Jebb's results, and he did not have some of the SNPs in the R-BY50723 branch, but his results alone were not enough to determine exactly where he belonged in the branch.
R-BY50723 is considered for now to be his "terminal" haplogroup, however this "terminal" haplogroup can change multiple times as more closely-related men take the Big Y test and provide a more precise placement on the tree.
In the branches below R-BY50723 there is only one SNP whose exact position in the haplotree is now known; it is SNP BY152878, and it has formed a new haplogroup called R-BY152878.
Here is the bottom of the above screen showing that Mr. Jebb is sharing branch R-BY50723 with one person from England.
 |
| Y-DNA matches in Big Y Block Tree |
You will notice in Mr. Jebb's branch that it says there is an average of 16 private variants between the two men in that branch.
Even though these two men are on the same branch of the haplotree, they are not considered to be a match because matches at FTDNA only include people who have no more than 30 total SNP differences which include private variants and named SNPs.
What are private variants?
In the Family Tree DNA Learning Center, FTDNA provides an explanation:
On the Big Y Block Tree, you will see blocks labeled Private Variants. Private Variants are one of the following;
- mutations that are not named nor are shared between any branch members.
- mutations that have not yet been validated and placed on the Haplotree.
It is important to note that Private Variants are filtered to only include SNP calls from regions of the Y chromosome that can be reliably mapped with NGS technology. For this reason, the block tree number might be lower than what you see in your personal Big Y Private Variants list.
Family Tree DNA used to list "Private Variants" as "Unnamed Variants" [See the "Examining variants with the Chromosome Browser" section of The Big Y could be the best DNA test ever!], but now most of them are named even though the official names do not appear on the list of private variants. Instead, private variants are listed by their position number on the Y chromosome.
As previously stated, the Block Tree indicates that there is an average of 16 private variants between the two men in haplogroup R-BY50723. We want to see the actual list of private variants for Mr. Jebb.
List of Private Variants
The private variants can be seen by clicking either the Matches link or the Results link in the Big Y section of the home page.
 |
| Big Y options |
You will then see a tab for Private Variants.
 |
Private Variants tab
|
I checked Mr. Jebb's Big Y results, and he had 22 private variants. The private variants are ones that were not seen in other men. They are listed below in three screens:
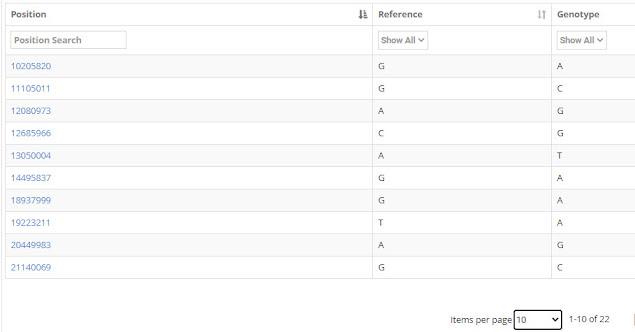 |
| Jebb Private Variants 1-10 |
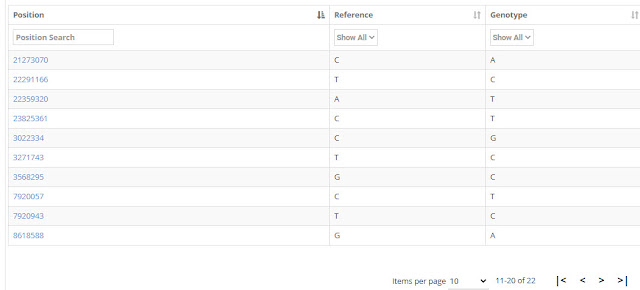 |
| Jebb Private Variants 11-20 |
 |
| Jebb Private Variants 21-22 |
With 22 private variants, Mr Jebb is remotely related to the other men on the Block Tree and is definitely not related to any of them within the genealogical time period.
Gibbs Big Y
The Gibbs Big Y results were not finished until September, but they were worth the wait. As suspected, he had one match:
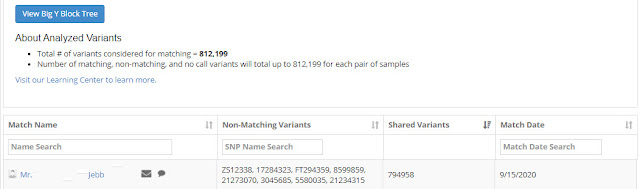 |
| Big Y match list |
In the above screen you can see the name of the match [I have erased his given name], an envelope for sending an email to the person, an icon for comments, the list of non-matching variants, the number of shared variants, and the date the match was discovered. We will examine some of these later.
In addition to accessing the Block Tree from your home page, you can click on the Blue link at the top of the above screen to "View Big Y Block Tree" and see your placement on the tree.
The Block Tree now looks quite different from the way it appeared before the Gibbs Big Y results were completed:
 |
| Big Y Block Tree with new match |
The Jebb-Gibbs branch of the tree is the white block on the right. The original R-BY50723 block of 26 SNPs (which was previously shown in white because it was considered to be Mr. Jebb's block) has been now been broken into two blocks called R-BY50725 (9 SNPs) and R-BY50723 (17 SNPs).
This is because Jebb and Gibbs both had the SNPs BY50725, BY50788, BY50811, BY50831, BY50848, BY73197, FT123034, FT123036, and FT94586, but they did not have the rest of the SNPs in block R-BY50723 which all of the other men have.
The SNPs in each block are listed in alphanumeric order because the order in which they occured is not yet known.
What is now known, however, is that all of the SNPs in block R-BY50725 occurred before the ones in block R-BY50723. Jebbs and Gibbs are now on their own new branch of the tree which is called R-FT368124.
Jebb and Gibbs common ancestors
Jebb and Gibbs share 20 SNPs in the block R-FT368124 that have not been seen before.
All of these SNPs represent an ancestor, but since a SNP did not occur at the birth of every male ancestor, these SNPs represent at least 20 generations of common ancestors.
In the left column of the Block Tree, you can see that Family Tree DNA estimates that their branch of the tree was formed about 33 SNP generations ago. If we use an average of 80-100 years per SNP generation, this branch is about 2640-3300 years old.
 |
| Estimated SNP Generations |
At the bottom of the branch is where we see the most recent common ancestor (MRCA) of Jebb and Gibbs.
The most recent common ancestor is when the two branches of Jebb and Gibbs separated. These two men have an average of 3 private variants since that time:
 |
| Private Variants on Block Tree |
You will notice that in the Family Tree DNA Block Tree the placement of more recent generations is far less reliable.
FTDNA seems to be placing the most recent common Jebb-Gibbs ancestor at about 13 SNP generations ago, but as you can see there is a wide variation of private and shared SNPs at the bottom of the tree.
Unless we have genealogical evidence of the most recent common ancestor, we can only broadly estimate the time to this ancestor by examining the private SNPs. This is because in some families Y-DNA mutates more than others, there may be uncertainty about the validity of some of the private variants, etc.
The date estimation becomes more accurate as more men take the Big Y test.
To find out more about the most recent common ancestor, we will need to examine the private variants that are not shared between the two men. This is the only part of the Big Y that may require work.
Before we look at the private variants, I will provide an explanation on how variants are identified.
How are variants identified?
Now we get to the difficult part. How do we know if a private variant or SNP is valid? How do we know whether we have more or fewer variants than FTDNA identified?
We must first understand how variants are determined. During the testing process, your DNA is not read in one continuous stretch. Instead, your DNA is broken into random fragments. The test reads these fragments from each end. These are called the forward read and the reverse read.
Some positions are read many more times than others. Some locations on the Y-chromosome can be read in one person's test and not in other.
After being read, the fragments are aligned to the reference sequence, and differences from the reference sequence are identified. The difference from the reference value is your "derived" value.
Unfortunately, not all of the reads may give the same result. One read may show the reference allele (for example a C) and another read may show a derived value (for example a G).
To be considered a high quality SNP by Family Tree DNA the position must usually be read at least ten times. The number of differing calls is then taken into account.
A position that was read a few times with different results will be considered to be a much less reliable variant than one that was read many times with a consistent result.
We will see how this works when we examine the Chromosome Browser in the next section.
Evaluating private mutations: Gibbs
One of the most interesting parts of the Big Y test is the list of private mutations. These are mutations that are not shared between the two men which means that the mutations should have occurred after the time of the most recent common ancestor.
As more and more men test their Y-DNA some of the private variants can eventually be identified with a specific ancestor.
If you go to the Big Y results, then to the Private variants tab, you will see a list of the mutations that were only seen in this test. Below, Mr. Gibbs has five private variants.
 |
| Gibbs Private Variants |
Click on any variant to see it displayed in the Y-Chromosome Browsing Tool (I refer to this tool as the Chromosome Browser). Here we are clicking on private variant 21234315.
In the Chromosome Browser you will see the various fragments being read during the test.
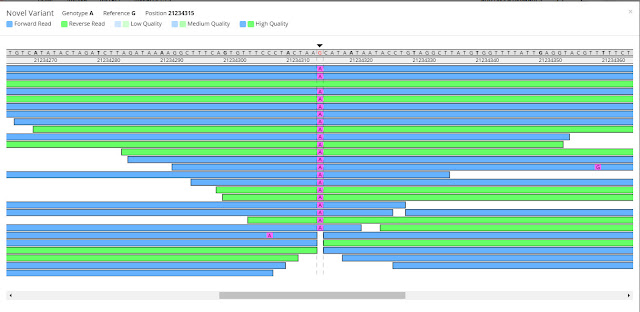 |
| Family Tree DNA Big Y Chromosome Browser |
In the above screen we are looking at the Gibbs private variant 21234315. The arrow at the top of the scan points to this position. It shows that the reference sequence had a G in this position, and then shows the various reads for the Gibbs test.
The fragments being read in the forward direction are indicated in blue; the ones read in the reverse direction are shown in green.
You can see where each scan started and stopped by scrolling across the bottom of the screen.
Notice on the third line that one of the reads showed a G in this position, but all of the rest showed an A. With 22 total reads, 21 showing A and only one showing G, this is considered to be a reliable private variant.
Although we know that private variants are named by Family Tree DNA, the name given to this position is not shown in the Chromosome Browser. However, we can find out the name given to this variant by going to ybrowse.org
 |
| Position 21234315 at ybrowse.org |
Position 21234315 has been identified as a SNP and has been given the name FT381499.
The names given to private variants will be important when we examine the variants of Jebb.
Examining private variants: Jebb
When we look at Mr. Jebb's Big Y results, and go to the Private Variants tab we see that he has only one private variant:
 |
| Big Y Private Variants |
Jebb and Gibbs share a common ancestor. After the time of the common Jebb/Gibbs ancestor, why did five mutations occur in the Gibbs descendants, and only one in the Jebb descendants?
It is known that mutations can occur for a variety of reasons including the age of the father (older fathers tend to pass down more mutations than younger ones), environmental factors, etc.
Was there a particular reason that the Gibbs line that left Ireland had more mutations than the Jebb line that remained, or did Mr. Jebb have some mutations that were not called by FTDNA? Were there some positions that were read in the Gibbs test but not in the Jebb test?
Unfortunately, FTDNA does not give us the tools to resolve any questionable variants that might have appeared in Mr. Jebb's test results.
Jebb's initial list of unnamed variants vs. final list
When Mr. Jebb's test was originally completed he had 22 private variants. After the results of Mr. Gibbs were completed and compared, the two men had 20 shared variants, and Jebb had only one private variant left. What happened to the missing variant?
We cannot use the Private Variants tab to answer this question because most of these are no longer private variants, and the Chromosome Browser does not show us the name given to any private variants. We must go to the Named Variants tab.
 |
| Big Y Named Variants |
I entered into the SNP search box the name of each SNP on the Block Tree that was only shared by Gibbs and Jebb. For each SNP click on the SNP name to see it in the Chromosome Browser.
 |
| Big Y Chromosome Browser |
The Chromosome Browser for Named Variants provides the SNP name and the position number. We now know that SNP FT381739 is at position 11105011 and was originally the unnamed variant of that name.
I went through all of the named variants and found that one of the previous unnamed variants did not appear. It is position 23825361.
You cannot use the FTDNA Chromosome Browser to for look this position.
We cannot search for it in the Named Variants because we do not know the name (if any) given to it.
We also cannot view it using the Private Variants section because this position now shows "Currently no results":
 |
| Search for Private Variant shows no results |
What happened to this variant that was previously on Jebb's list of Private Variants? Again, we can search for any position using YBrowse:
 |
| Unnamed Y-chromosome position in YBrowse |
This position was not given a name.
At FTDNA you can only examine the private variants that appear on your list. So there is no way at FTDNA to see why that position was not named.
In both Jebb and Gibbs, if I search for position 23825361 I see the message "Currently no results." I cannot see this position in the Chromosome Browser, so there is no way at FTDNA to see if this position was read at all in either test or why this position was not named as a SNP or was not on Jebb's list of private variants.
There is only one way to find out about this variant, and that is to examine either the VCF file or, even better, the BAM file that are available for download.
VCF and BAM data files
First, let's acknowledge that few people have expertise, or even interest, in examining raw data files. But you don't have to do any analysis yourself. There are others who will do it for you. We will see some of those options below.
From either the Big Y Results section or the Big Y Matches section you will see links to Download VCF or Buy Raw Data (the BAM file).
The VCF file is a filtered data file that can be quite useful, but the BAM file is the one that is needed to identify all possible private variants.
 |
| Download Raw Data |
Downloading the VCF file is free. Unfortunately, the BAM file is under "Buy Raw Data" and FTDNA is now charging $100 to download this file.
Here's some positive news about the $100 fee: At the time this fee was imposed FTDNA reduced the price of the Big Y by the same amount, so you might consider that you are now paying for the Big Y in two stages.
Once you download either the VCF or BAM, let's see options for evaluating them.
Transfer the results to YFull
Transferring your results to Yfull.com is, by far, your best option. Yfull will accept either the VCF or BAM file.
If you choose to submit the VCF file, you can later submit the BAM file at no additional charge.
Even without the BAM file, the VCF file can be very useful, and you will still find out much more about your Y-DNA and be able to use YFull tools with this file.
For much more about the many benefits of YFull and how to use them see the following two blog posts:
The Gibbs VCF file has been transferred to YFull, so even though Jebb's file is not there yet, we can still find out more about Jebb's missing private variant.
YFull uses a different name for "Private Variants." They are called "Novel SNPs." YFull will separate Novel SNPs from a VCF file into three categories:
 |
| YFull Novel SNPs identified from VCF file |
YFull separates Novel SNPs from BAM files into five categories. For example, in the image below, these "One reading" variants could only be determined from a BAM file.
After examining the BAM file YFull will show more variants than Family Tree DNA does.
 |
| YFull Novel SNPs identified from BAM file |
One of the advantages of YFull is that you can search for any variant in your file. Click Browse Raw Data in the YFull menu:
 |
| Browse Raw Data |
Although I could not find out why Jebb's private variant 23825361 is no longer on his list and could not see this position in the Chromosome Browser for Gibbs at FTDNA, I can see this position in the Gibbs file at YFull.
 |
| Browse Raw Data at YFull |
In the above image you can see that at position 23825361 the Reference sample has a C (Reference allele). This position was read 129 times in the Big Y test for Gibbs.
In the Position Data row, it shows that the Gibbs results had a read of "T" 66 times at this position, and had a read of "C" 63 times. The reads at this position are unreliable, so during the manual review Family Tree removed 23825361 from Jebb's list of private variants.
Summary of Jebb and Gibbs Big Y results
Here's what we now know for certain. The Canadian immigrant William Gibbs and the Jebbs family of Billis, County Monaghan, share a common ancestor.
What we haven't proven yet is the name of the common ancestor. We can find out much more testing other Jebb descendants and by examining the shared and private variants of these men.
So what are the next steps for Jebb and Gibbs?
In the next section I will provide a list of suggestions for all Y-DNA testers no matter what level of Y-DNA testing you have ordered.
What to do after getting your Y-DNA results
There is always more to learn from our DNA. It's the gift that keeps giving. We must let others know about our discoveries and preserve this information for future generations. Getting the results is just the beginning!
Although there are many more things we can do beyond what is listed below, here is a list of what I consider to be essential actions if you tested Y-DNA at Family Tree DNA. The list may vary if you tested at another company.
If you want to make great matches with Y-DNA follow all of the steps below.
At Family Tree DNA:
1. If you already have a FTDNA account, click Add ons & Upgrades at the top of the screen. If you have not ordered a Y-DNA test, you will click the Add ons tab. If you want to upgrade your STR test click the Upgrades tab.
 |
Add Ons and Upgrades
|
2. Add information about your earliest known paternal ancestor. Hover the mouse over your name, then click Account Settings.
 |
| FTDNA Account Settings |
From the Account Settings page, click Genealogy, then Earliest Known Ancestors.
 |
| Provide name, date, and place for earliest known paternal ancestor |
Enter the name of your earliest-known direct paternal ancestor.
Even though the field says to enter birth and death dates, it is much better to enter a year of birth and the approximate birth location. This is what will appear on match lists and in FTDNA projects.
Enter a precise location using the location field below the ancestor's name. This is location that will appear in the Matches Maps section of your Y-STR results.
3. Review your Privacy Settings. Click the Privacy & Sharing tab. Review all privacy settings, and make sure you have opted into matching.
 |
| Verify FTDNA account settings |
Select your Y-DNA match level. In most cases you will want to select All Levels.
 |
| Select Y-DNA match level at FTDNA |
4. Add a family tree to your account. Family trees at FTDNA are great for obtaining basic ancestral information, but the trees do not include sources.
So, in addition to adding a family tree to their FTDNA account, some people put links to other online trees in the "About Me" section of their profile. To do this, go to Account Settings>Account Information>My Personal Story.
Many people may not look at your profile, so be sure to add a family tree to your FTDNA account. You can even link DNA matches to your family tree.
Click myTREE at the top of your home screen.
 |
| View your family tree at FTDNA |
You will then be given the opportunity to create your tree.
 |
Create your family tree at FTDNA
|
You can simply create a tree that contains only your paternal line. This is not your best option, however, because you will want a more complete family tree for Family Finder results, mtDNA tests, and any other tests that may be offered in the future.
If you already have a family tree, click UPLOAD GEDCOM. You can create a GEDCOM file from your genealogy software program or from many online family trees.
For example, you can download your family tree from Ancestry.com and upload it as a GEDCOM to your Family Tree DNA account. To download your tree from Ancestry, log into your Ancestry.com account. Click the name of your tree, then click Tree Settings.
 |
| Ancestry.com tree settings |
Find the "Manage your tree" section on the right of the next screen, then click Export tree.
 |
| Export family tree as GEDCOM file |
Ancestry will send you an email verifying that you are the account owner, then you will be able to download the GEDCOM and upload it to your Family Tree DNA account.
5. Join a FTDNA surname project. Go to myPROJECTS at the top of your FTDNA home page, then click Join A Project.
 |
Join a Project
|
Go to the Surname Projects section then click the first letter of your surname.
 |
| Surname Projects at FTDNA |
There was not a Jebb surname project, but there was a Gibbs project. The project administrator has now added Jebb and Jebbs to the list of surnames for the project, and now both men appear together where we can compare the 111 STRs.
 |
| STR results in Gibbs Surname Project |
6. Join a FTDNA haplogroup project for your Y-DNA haplogroup. One of the most important reasons for joining a haplogroup project is to extend your paternal line by finding men who are slightly more related to you than your closest surname matches.
First, find your haplogroup. If you have only tested STRs you will be assigned to an ancient haplogroup. One of the most common is R-M269. You can find your haplogroup on the left side of the screen in the Badges section.
 |
| Haplogroup Badge |
You can also find your haplogroup in the Y-DNA section of your home page.
 |
| Haplogroup recorded in Y-DNA section of FTDNA account |
At the top of your FTDNA home page, click myPROJECTS, then Join A Project. (You can also find this option on the left side of your home page.)
 |
Join a Project
|
Find the Y-DNA Haplogroup Projects, then click the first letter of your Y-DNA haplogroup.
 |
| Y-DNA haplogroup projects at FTDNA |
The Y-DNA haplogroup list can be confusing because some projects were formed when Family Tree DNA used an earlier system of naming haplogroups. For example, there is no haplogroup project named R-M269.
 |
| Haplogroups listed at top of Big Y Block Tree |
The haplogroup project for R-M269 is called "R _R1b ALL Subclades":
 |
| R1b haplogroup project at FTDNA |
If you have taken the Big Y test, there is likely not a haplogroup project for your specific terminal haplogroup which may only include a few people. However, your terminal haplogroup is a subclade of haplogroups formed earlier, and you can join a project for one or more of those haplogroups.
You can find a list of your haplogroup lineage, from ancient to modern, at the top of your Big Y Block Tree.
 |
Y-DNA haplogroups listed at top of Big Y Block Tree
|
Again, go to Join A Project, and in the Y-DNA Haplogroup Projects click the name of the project you wish to join. For example, Haplogroup R-FT368124 assigned to Jebb and Gibbs is a subclade of R-U106, so they should both join that project.
 |
| Join haplogroup project |
On the next page click Join. Repeat this process to join multiple haplogroup projects. You can join as many upstream projects as you wish.
7. Update your Group Project Administrator Access. As you have done before, click on Account Settings, but this time click Project Preferences. Click the pencil icon next to the name of your project. In most cases, it is recommended to give all project administrators limited access.
 |
| Group Project Administrator Access |
8. Download your Y-STR results. You will use this file to upload to mitoYDNA and, for Big Y tests, to update your results at YFull [both described below]. Go to the Y-DNA section of your FTDNA home page, and click Y-STR Results.
 |
| Click on Y-STR Results |
Scroll to the bottom of the results and click Download CSV.
 |
| Download CSV file of STRs from FTDNA |
At MitoYDNA:
MitoYDNA replaces the databases YSearch and Mitosearch that had been created by Family Tree DNA but are now removed. MitoYDNA was created by private citizens, and not by a company, so its database should remain. You will use this database to compare and preserve your Y-STR results.
Go to mitoYDNA.org and register. Log into our account, and click Kits at the top of the screen.
 |
| Create new Y-DNA kit |
Click Create a new kit, then fill out the information on the next page. At the bottom of the screen click "Choose File" to upload the CSV file that you downloaded from Family Tree DNA.
 |
| Choose CSV file |
You also have the option to click "Manually Entered" to individually enter your mutations. Once you have created your file you may want to take a screen shot of it and upload it to your ancestor's profile in your online family trees.
If you have taken a Big Y test
At Family Tree DNA:
9. Go to your Big Y results, and download your VCF file. You can use this to submit to YFull and the Y-DNA Data Warehouse (see below). This is a different file from the CSV file of STRs that you previously downloaded.
 |
| Download VCF File from FTDNA |
10. I strongly recommend that you also click Buy Raw Data and preserve your BAM file.
At YFull:
1. Go to yfull.com and place your order.
 |
| Order Y-DNA analysis at YFull |
Notice in the above image that uploading the VCF file does not include STR matching. I will show you how to get around this issue.
Fill out the order information, and load your VCF file on the next screen.
 |
| Upload VCF file to YFull |
Once you have uploaded Y-DNA results, you can add mtDNA results at no additional charge. Log into your YFull account, and in the menu on the left click Upload mtDNA.
 |
| Upload mtDNA to your YFull account |
Then upload a FASTA file of your mtDNA results.
You can also upload your BAM file of Big Y results at no additional charge.
2. Enter information about your most distant known paternal ancestor. Click Settings at the upper right of your Home page.
 |
Update account settings
|
Click Most Distant Ancestor.
 |
| Enter Most Distant Ancestor and Place of Origin |
Enter your ancestor's name along with the approximate year and place where he was born. Be as specific as you can. Again, in the screenshot below, the example below the ancestor field shows the name John Johnson with dates of birth and death. This is not very helpful. We need a name, date, and PLACE.
 |
| YFull: Most Distant Ancestor |
Next, click Country of Origin. Enter not only the country but also the region, if known.
 |
Add Country and Region of Paternal Origin
|
3. After your results are ready, upload your CSV file of STRs to your YFull account. YFull can extract STRs from your BAM file, but they cannot extract them from the VCF file.
 |
| STR matches at YFull |
Please note, however, the neither the BAM nor the VCF contains the complete list of 111 STRs that can only be obtained from the CSV file. To submit your CSV file of STRs, go to the STRs section of the YFull menu, and click Upload STRs.
 |
| Upload CSV file of STRs |
You will now be able to compare all STRs (not just the first 111) in YFull groups.
4. Join a group if one exists for your haplogroup. YFull "groups" are similar to FTDNA "projects".
In the menu on the left, scroll down to Groups Y.
 |
| Y-DNA Groups at YFull |
On the next page click the name of the group you wish to join, then click Join request.
 |
| Submit Join Request |
If there is no Y-DNA group for your haplogroup or for your surname you can easily start one yourself. Send a message to YFull stating something like, "I would like to start a Y-DNA group for the surname Jebb." Your new group will be quickly formed. You will periodically receive an email when a new person wants to join your group. All results within the group are automatically sorted, but you can create your own subgroups if you wish. For example, subgroups were created in the R-U106 group, and Mr. Gibbs is in the subgroup "Z2265+ Z381- Z18-".
 |
| YFull Group with subgroups |
The complete list of STRs can now be compared in a YFull group. For example, even though Gibbs submitted a VCF fie and has no matches in the STR matches section of his YFull account, he can make STR matches in YFull groups because he also submitted a CSV file. In the screenshot below, the STR results of Mr. Gibbs are in the fifth row from the top:
 |
| Comparison of STRs in YFull group |
At Y-DNA Data Warehouse:
The primary use of the Y-DNA Data Warehouse has been to submit VCF files for inclusion in Alex Williamson's The Big Tree for haplogroup R-P312 and subclades. The Warehouse has recently been expanded to include all haplogroups, although it is not yet known if the other haplogroups will be displayed online. This will be a subject for a future blog post. In the meantime, go to Y-DNA Data Warehouse and upload your VCF file. This is especially important if your haplogroup is within haplogroup R-P312 so that you can get into The Big Tree. See an example of a subclade in The Big Tree here: R-A9871.
Recruit other men for Y-DNA testing
Recruit other male descendants of your ancestral line to test their Y-DNA. For example in the case of Jebb and Gibbs, we do not yet know their common ancestor but the DNA results of other Jebb/Gibbs men with known family trees can provide the answer. I will definitely try to recruit another Jebb with ancestors from County Monaghan.
a. Contact your Y-DNA matches. They likely share your interest and may help recruit other men with your surname to test their Y-DNA.
b. From your surname or haplogroup project at Family Tree DNA, post in the group's Activity Feed (if the group is using that feature).
c. Join an online forum for your haplogroup.
d. Recruit other testers in surname societies and their newsletters or online forums.
e. Post links or screenshots of STR results in online family trees. People may contact you regarding these results.
f. Communicate with people who share your common ancestor
Summary
The basic genetic outline for the Jebb and Gibbs families has been provided by the Big Y results of two men. All future descendants of their Irish ancestors will provide a more complete picture. The best results will be obtained by following all of the steps above.
For yourself. please complete as many of the steps above as you can. Then after you're finished contact your Y-DNA matches, and encourage them as well. If more people preserve and compare their results as outlined above we could be seeing some amazing results in a relatively short time.
______________________________________________________
Disclosure
Links to Family Tree DNA appear in the sidebar. I receive a small contribution if you make a purchase, but clicking through the link does not affect the price you pay.








































































5 comments:
Great write up of some otherwise tedious steps! As many of us have learned by fumbling around with these tools and results over the years one has to put in some additional work to make the interpretation of the results meaningful. This example is notable for helping in the most general cases all the way down to specific Y-SNP results on YFull.com. I recommend this blog post to all surname and haplogroup projects!
My compliments on a depth of analysis from many sources that is rarely achieved. A good read for any serious dna and family tree analyst.
This was so informative. I, too, tested for Y and Mt at Ancestry, Sorensen, and NG Genographic in the old days. Since then, all FTDNA tests including Big Y-700, LivingDNA, Geno 2.0, and Dante Labs WGS. I've joined many different
projects, also submitted to Gedmatch, joined many different DNA Facebook groups. After 20+ years, I know a lot, but still feel like I know nothing compared to others. I have still not been able to share my Dante WGS info with FTDNA for analysis because of the file size. Since I live in Houston, maybe I can hand deliver a flash drive.
Fantastic, instructive and very informative, thank you.
One thing that I would like to add that I recently discovered with my nearest Big-Y700 match results: The list of "Private Variants" MAY INCLUDE variants found in other nearby Y700 match "Private Variants" lists. This can create a huge difference between the actual and reported difference and time estimate to common ancestor.
My nearest Y700 match had 10 Private Variants before I took the Y700 upgrade.
After, he still had 10 Private Variants.
Only after we compared our lists, we found that of his 10, 4 are also on my list of 12 Private Variants.
And so, instead of 10 mutations, his common ancestor with me is only 6 mutations back.
That is a huge, nominally 400 years difference!
Of my 12 Private Variants, we know now that 4 of them predate that common ancestor. Of the 8 remaining, 1 is shared with my nearest Y500 match and so is about 400 years back. Of the 7 remaining, 3 are Y700 new and the of the other 4, two (2) occurred with my own father!
On the Block Tree, rather than having an average of 6 mutations (12 from me, 0 from my nearest Y500 match), we know this is off by 2 mutations due to the 12 actually being 8.
Anyway, be careful to not assume that "Private Variants" mean that no other Y700 testers in your local tree have those variants. They could easily share them but that FTDNA does not know where to place the common variants if the two individuals do not share the samem final haplogroup. That is what happened in my case.
Thank you for posting that comment. It might explain a big difference between the paper trail genealogy (MRCA 1700's) and the FTDNA estimate (MRCA 1500's) in my own line & that of a distant cousin.
If you could explain how you managed to compare private variants with that other match, it would be much appreciated!
Regards from Byron Wade.
Post a Comment