For Y-DNA testing, I have seen a lot of questions about how to evaluate Next-Generation Sequencing (NGS) tests such as Family Tree DNA's Big Y test, and similar tests at Full Genomes Corp, YSEQ, and extracted Y-DNA results of full genome tests from companies such as Dante Labs and Nebula Genomics.
If your Y-DNA company gave you good results, why would you want to download your results and upload them to a third-party site such as YFull?
I previously wrote a post about the advantages of YFull, but that post was written when I took my original Big Y test. Please review What are the benefits of YFull?
I previously wrote a post about the advantages of YFull, but that post was written when I took my original Big Y test. Please review What are the benefits of YFull?
The benefits are much more extensive now that YFull has added new tools.
So, what are the advantages of submitting to YFull?
YFull is not a DNA testing company. YFull is an analysis and comparison service for Y-DNA Next Generation Sequencing and full mitochondrial DNA sequences.
What is YFull?
YFull is not a DNA testing company. YFull is an analysis and comparison service for Y-DNA Next Generation Sequencing and full mitochondrial DNA sequences.
These interpretation and comparison services are more comprehensive than those found at any DNA testing company.
In this post we will discuss Y-DNA only. Since most people at the present time have tested with Family Tree DNA, let's focus on a portion of the Big Y-700 results from Family Tree DNA and compare these to the YFull evaluation of the same results.
We will examine the Private Variants from a Big Y-700 kit at Family Tree DNA.
Y-DNA analysis and comparison
In this post we will discuss Y-DNA only. Since most people at the present time have tested with Family Tree DNA, let's focus on a portion of the Big Y-700 results from Family Tree DNA and compare these to the YFull evaluation of the same results.
We will examine the Private Variants from a Big Y-700 kit at Family Tree DNA.
Private Variants are those that have supposedly only been seen in your kit. Once they have been seen in more than one kit, you will find them in the Named Variants.
However, note that I said "supposedly." Sometimes positions appear in your Private Variants list that have been seen in other kits, but FTDNA hasn't yet discovered this. This will especially be the case if your results have just arrived and haven't been fully evaluated.
Log into the Family Tree DNA account and go to Big Y Results.
Log into the Family Tree DNA account and go to Big Y Results.
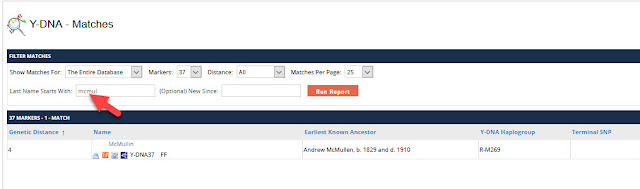
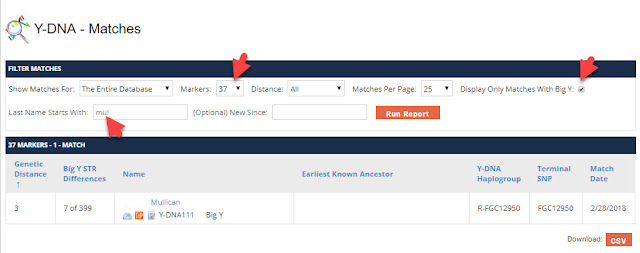
There are three tabs for Named Variants, Private Variants, and Matching. Here are the first ten Private Variants for a Big Y-700 kit. There are a total of 19 private variants, but only the first ten are displayed below. They are listed by their hg38 position number:
 |
| Big Y-700 Private Variants |
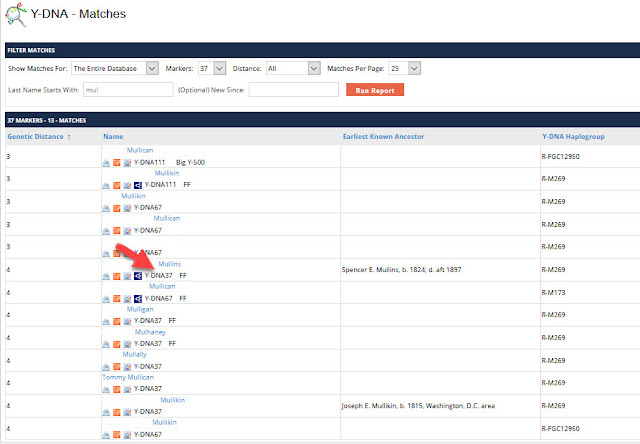
Let's look at the first SNP on the list, position 10053444. If we click on the blue link for that position, you will be taken to Family Tree DNA's chromosome browser and see the following:
 |
| Big Y chromosome browser |
This SNP is considered by FTDNA to be high quality. The Reference Sequence has a G at this position, but this kit had a C. The chromosome browser shows us how many times the position has been read, but we have to count each line.
No further information is available. Now let's see what we can discover about this same position at YFull.
In YFull, "Private variants" are called "Novel SNPs." Here again, the SNPs may not truly be novel. Other companies may have found these variants, but YFull hasn't discovered them yet. To see a list of your novel SNPs, view the menu at the left side of the screen, and click on Novel SNPs.
At YFull, SNPs are categorized by quality and divided into separate tabs. The tabs are Best qual [best quality], Acceptable qual, Ambiguous qual, Low qual, One reading, and Indels.
Below is the Best qual tab. You will see the SNPs listed by both their hg19 and hg38 positions. Notice that at FTDNA we only saw the hg38 position number. To see if we can find anything new about the position we examined at FTDNA, find 10053444 on the Best qual screen.
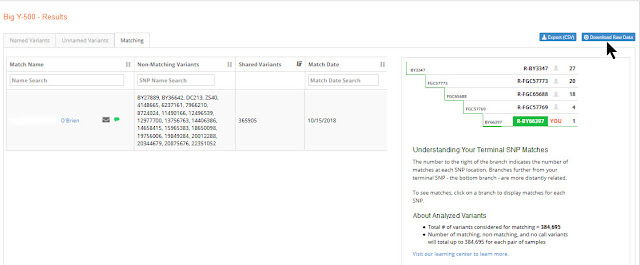
You can see that 10053444 has been named FGC65817 by Full Genomes Corp, but we did not see that at FTDNA. The red check mark on the line means that this SNP is available for verification by Sanger testing at YSEQ. This is another piece of very useful information, as we will see later.
Now click View BAM on the right side of the screen to see several versions of the BAM viewer. This one is similar to the one we saw at FTDNA:
We can see even more by clicking the yellow magnifier icon on the left side of the screen:
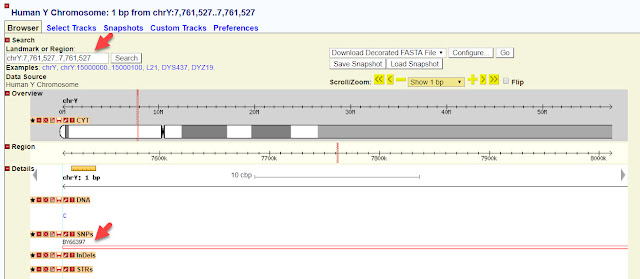
We will then see this information:
The above screen tells us the kit number and haplogroup [on the first line, but much of it is erased in this image] and the Y-chromosome position numbers in hg19 and hg38. The red arrow next to the hg38 position shows that we are looking at a test that was aligned to hg38. This position was read 22 times in the Big Y test, and all the reads showed a C instead of the G found in the Reference Sequence. We also see that the SNP has been named FGC65817, it is available for testing at YSEQ, and it is listed in YBrowse.
The right side of the above image was cropped to make it more readable. However, another useful part of this screen is that you can not only see the position number, but you can see where this position is situated on the Y chromosome. This can help you determine how reliable this SNP may be.
Novel SNPs at YFull
In YFull, "Private variants" are called "Novel SNPs." Here again, the SNPs may not truly be novel. Other companies may have found these variants, but YFull hasn't discovered them yet. To see a list of your novel SNPs, view the menu at the left side of the screen, and click on Novel SNPs.
 |
| Click Novel SNPs |
At YFull, SNPs are categorized by quality and divided into separate tabs. The tabs are Best qual [best quality], Acceptable qual, Ambiguous qual, Low qual, One reading, and Indels.
Below is the Best qual tab. You will see the SNPs listed by both their hg19 and hg38 positions. Notice that at FTDNA we only saw the hg38 position number. To see if we can find anything new about the position we examined at FTDNA, find 10053444 on the Best qual screen.
 |
| List of Novel SNPs at YFull |
You can see that 10053444 has been named FGC65817 by Full Genomes Corp, but we did not see that at FTDNA. The red check mark on the line means that this SNP is available for verification by Sanger testing at YSEQ. This is another piece of very useful information, as we will see later.
Now click View BAM on the right side of the screen to see several versions of the BAM viewer. This one is similar to the one we saw at FTDNA:
 |
| One version of YFull chromosome browser |
We can see even more by clicking the yellow magnifier icon on the left side of the screen:
 |
| View position in BAM |
We will then see this information:
 |
| SNP information |
The above screen tells us the kit number and haplogroup [on the first line, but much of it is erased in this image] and the Y-chromosome position numbers in hg19 and hg38. The red arrow next to the hg38 position shows that we are looking at a test that was aligned to hg38. This position was read 22 times in the Big Y test, and all the reads showed a C instead of the G found in the Reference Sequence. We also see that the SNP has been named FGC65817, it is available for testing at YSEQ, and it is listed in YBrowse.
The right side of the above image was cropped to make it more readable. However, another useful part of this screen is that you can not only see the position number, but you can see where this position is situated on the Y chromosome. This can help you determine how reliable this SNP may be.
As shown below, position 10053444 is found in the Yp11.2 combBED region:
Notice that the fourth position below has been given the SNP name FT86640. SNPs beginning with FT are ones that were discovered by Family Tree DNA from the Big Y-700 test. Since it was not discovered in the previous Big Y test, let's see if this could be a valid SNP. Click the magnifier.
If we click the Ambiguous quality tab, we see the following. Let's examine the first listed position to see why it's on the ambiguous list. Again, click the magnifier on the left.
On the next screen we see that this SNP was only read two times, so it's a less reliable SNP.
Family Tree DNA does not show this position in its list of Private Variants because it was only read two times. YFull considers the above SNP to be ambiguous for the same reason, but they do list it.
| Y-chromosome regions |
Notice that the fourth position below has been given the SNP name FT86640. SNPs beginning with FT are ones that were discovered by Family Tree DNA from the Big Y-700 test. Since it was not discovered in the previous Big Y test, let's see if this could be a valid SNP. Click the magnifier.
 |
| YFull Novel SNPs |
This SNP had consistent results for 39 reads, so that's a good sign.
 |
| Search in BAM file |
If we click the Ambiguous quality tab, we see the following. Let's examine the first listed position to see why it's on the ambiguous list. Again, click the magnifier on the left.
 |
| Evaluate an ambiguous SNP |
On the next screen we see that this SNP was only read two times, so it's a less reliable SNP.
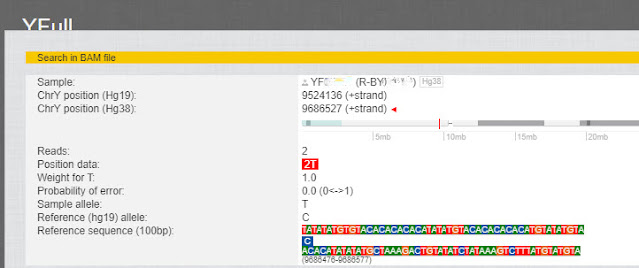 |
| A SNP with two reads |
Family Tree DNA does not show this position in its list of Private Variants because it was only read two times. YFull considers the above SNP to be ambiguous for the same reason, but they do list it.
If you want to know if any SNP is a valid one, you can verify it by ordering Sanger Sequencing at YSEQ for this position (along with any other doubtful SNPs). YFull does not indicate that the 9686527 SNP is available for testing at YSEQ, so go to yseq.net, and check to see if it's been added to their list of SNPs. If not, submit the position to YSEQ's Wish A SNP:
On the next page, you will see full instructions for making your SNP available for testing. Notice that the price is only one dollar.
If this SNP is in a region that can be reliably tested, you will receive an email from YSEQ when your SNP is available.
 |
| YSEQ Wish a SNP |
On the next page, you will see full instructions for making your SNP available for testing. Notice that the price is only one dollar.
 |
| YSEQ Wish a SNP order |
If this SNP is in a region that can be reliably tested, you will receive an email from YSEQ when your SNP is available.
Because 9686527 was only read twice in the Big Y test, it is a questionable SNP, but once it's available for testing you can submit a DNA sample to YSEQ to verify that you actually have a novel SNP at this position.
The ability to verify questionable SNPs is very important when comparing your results to someone else.
Now go to YFull.com and click Order Now:
The cost is $49, but you will not be charged until the results are ready.
How do I download my Big Y results and submit them to YFull?
Log into your FTDNA account, and go to Big Y Results. Click the blue Download Raw Data link at the upper right of the screen:
 |
| Download Raw Data |
If you haven't already done so, you will first have to request the BAM file. In a few days your BAM file will be ready. When it is, click Share BAM. then copy the link that appears.
 |
| Share BAM file |
Now go to YFull.com and click Order Now:
 |
| Order YFull interpretation |
The cost is $49, but you will not be charged until the results are ready.
If you have previously submitted another kit for the same person (for example, you previously submitted Big Y-500, and now you're submitting Big Y-700), add a comment to your order that "This is the same kit as [YFxxxxx]" and insert your old YFull ID number. You will get a new ID number for your Big Y-700 results, and it will be cross-referenced to your old ID number in the YFull tree.
If you have ordered a mtDNA Full Sequence test for the same person, those results can also be uploaded at no additional charge.
Comparing STRs
To be sure you have the most complete results in your YFull account, you will want to upload a STR file as well as your BAM file. This is because the STRs are not as reliable from NGS tests as they are from Sanger testing.
Comparing STRs
To be sure you have the most complete results in your YFull account, you will want to upload a STR file as well as your BAM file. This is because the STRs are not as reliable from NGS tests as they are from Sanger testing.
Family Tree DNA does Sanger testing to get the first 111 STR markers, but the BAM file you uploaded does not include the Sanger-tested results.
Be sure to upload not only your BAM file (as shown above), but also upload a separate STR file. No matter what company you used for your NGS test, you can order a STR test from Family Tree DNA or from YSEQ.
If you ordered a STR test from FTDNA (it was included in any Big Y-700 results), log into your FTDNA account and go to your Y-STR Results page:
Scroll all the way to the bottom of the page, and click the orange CSV button at the bottom right of the screen:
In your YFull account, click the Upload STRs link.
Depending on whether you received your STRs from YSEQ or FTDNA, on the next page either click Upload STRs - FTDNA or Upload STRs -YSEQ:
Notice above that there are two kits in the account. No CSV file of STRs has yet been uploaded for the first kit on the list. The FTDNA CSV file for the second kit has been loaded.
If you ordered a STR test from FTDNA (it was included in any Big Y-700 results), log into your FTDNA account and go to your Y-STR Results page:
 |
| FTDNA Y-STR Results |
Scroll all the way to the bottom of the page, and click the orange CSV button at the bottom right of the screen:
 |
| Download CSV file of STRs |
In your YFull account, click the Upload STRs link.
 |
| Upload STRs |
Depending on whether you received your STRs from YSEQ or FTDNA, on the next page either click Upload STRs - FTDNA or Upload STRs -YSEQ:
 |
| Upload FTDNA or YSEQ STRs |
Notice above that there are two kits in the account. No CSV file of STRs has yet been uploaded for the first kit on the list. The FTDNA CSV file for the second kit has been loaded.
The green check mark means that the CSV file passed the quality check. The red X is an option to delete the file.
The Re-upload link is so that you can upload another CSV file if you get any additional STR results for the same kit from FTDNA.
You can also upload a STR file from YSEQ for the same account. The STR uploads are free.
With your STR CSV file uploaded, you will get better results from YFull Groups. "Groups" at YFull are similar to "Projects" at FTDNA. You can submit a request to form a new group by sending an email to YFull.
To join a YFull group, go to the Others section, and click Groups Y.
On the next page, find the group of interest and click the name of that group. You will be taken to the Group page where you can submit a request.
Once you have joined a group, you can see the group results.
YFull Groups
With your STR CSV file uploaded, you will get better results from YFull Groups. "Groups" at YFull are similar to "Projects" at FTDNA. You can submit a request to form a new group by sending an email to YFull.
Joining a Group
To join a YFull group, go to the Others section, and click Groups Y.
 |
| YFull Y-DNA Groups |
On the next page, find the group of interest and click the name of that group. You will be taken to the Group page where you can submit a request.
 |
| Request to join a group |
The Join request form looks like this:
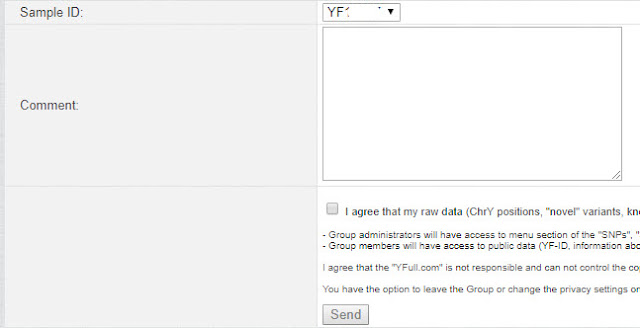 |
| Request form to join a YFull Group |
Select the correct sample ID [if you have more than one sample in your account], then submit a comment. You do not need to enter a comment. Check the "I agree" box, and click Send. You will receive an email as soon as your request is approved.
Advantages of Groups
Below are the first lines of the results from the R-L21 group. Notice that only twelve markers are displayed. This is the default, but you can display 12, 25, 37, 67, 111 or ALL markers.
Family Tree DNA only displays the first 111 markers in projects because the STRs from the Big Y tests are less reliable than the first 111 markers tested by Sanger testing.
Notice above that some of the STRs are missing or questionable in the test results. This means that the NGS test returned reliable results for some positions and not others.
 |
| YFull R-L21 STR results |
Notice above that some of the STRs are missing or questionable in the test results. This means that the NGS test returned reliable results for some positions and not others.
However, if these were results from Family Tree DNA all of the first twelve markers should have solid results because FTDNA does Sanger testing on the first 111 markers. In the above screen, the people who have missing or questionable results loaded their BAM file, but they did not load their STR file.
We can display all STRs by clicking the All view button as shown above.
The results below are some of the additional STRs received from the Big Y-700 test that are not compared at Family Tree DNA.
In the above screen we can see that the NGS tests do not return all STRs for all people.
We can display all STRs by clicking the All view button as shown above.
The results below are some of the additional STRs received from the Big Y-700 test that are not compared at Family Tree DNA.
 |
| YFull Comparison of all STRs |
In the above screen we can see that the NGS tests do not return all STRs for all people.
This is one reason why FTDNA does not compare them, but if so, what is the benefit of showing them at YFull?
One reason is that if you discover missing STRs, you may be able to order these from YSEQ and have them added to your account. We will examine this procedure later in this post.
Although FTDNA projects are generally much larger than YFull groups, YFull groups have a few major advantages.
Although FTDNA projects are generally much larger than YFull groups, YFull groups have a few major advantages.
The first advantage is your ability to contact anyone in the group whose results interest you. [For example, let's say that you see that someone else appears to be an STR match to you, but his test did not show results for many STRs that you have. Has he uploaded his STR file? Would the two of you be willing to do Sanger testing for any STRs that you both agree are particularly important?]
Simply click the PM (Private Message) envelope icon next to the kit number in the group results. A message screen will pop up:
Notice that you are sending a message to Kit YF06227 who is recipient 6227. We do not know this person's name or email address.
 |
| Sending a private message in YFull |
Notice that you are sending a message to Kit YF06227 who is recipient 6227. We do not know this person's name or email address.
However, once you send the message it will appear in that person's YFull account, and they will have the option to respond.
A second advantage is that YFull groups have results from not only Family Tree DNA, but also from other companies. This will be increasingly important as the price of Full Genome testing continues to come down.
A second advantage is that YFull groups have results from not only Family Tree DNA, but also from other companies. This will be increasingly important as the price of Full Genome testing continues to come down.
More and more people are extracting Y-DNA results from Full Genome tests and uploading the results to YFull.
In addition, YFull group administrators can add results from scientific studies. See the "Add science sample" below.
A third benefit of YFull groups is the ability to search for SNPs to find everyone in the project who might share your SNP of interest.
You can search by SNP name using the Y-Results tab or by position number using the Y-Browser tab.
 |
| Add science sample to YFull Group |
A third benefit of YFull groups is the ability to search for SNPs to find everyone in the project who might share your SNP of interest.
 |
| Search for SNPs |
You can search by SNP name using the Y-Results tab or by position number using the Y-Browser tab.
Although the Group's scientific samples may not show in the STR table (because the STRs were not included in the scientific report), they will show up in the SNP search.
Searching by "View Y-SNPs", people who have the SNP we're looking for will show up in the results list with a + sign like this:
Of course, if any SNP is one of particular interest, we could send a Private Message to another person who has this SNP.
The results will be less likely to show positive results when searching for something like an ambiguous novel SNP. Using the Y-Browser, we searched by position. The results will show every sample in the Group.
Searching by "View Y-SNPs", people who have the SNP we're looking for will show up in the results list with a + sign like this:
 |
| Positive SNPs |
Of course, if any SNP is one of particular interest, we could send a Private Message to another person who has this SNP.
The results will be less likely to show positive results when searching for something like an ambiguous novel SNP. Using the Y-Browser, we searched by position. The results will show every sample in the Group.
Only the first three results are shown in the image below:
These results show that the Reference Sequence had an A at position 14239619.
 |
| Search by Position number |
These results show that the Reference Sequence had an A at position 14239619.
The first ID had a N which means this position had no reads in the Big Y test.
The second ID had an Error. The error is because the position had only two reads, and they were both T.
The third ID is a scientific sample. The sample had an A in this position. If we hover the mouse over the A, we can see that this position was read five times in the test.
The ability in Groups to search by SNPs is another tool that can verify SNPs and find variants that can identify recent lineages.
What if I have taken more than one test?
One really great thing about YFull is the ability to load several test results for the same person. Your previous results do not disappear.
For example, I ordered a Big Y test for a kit in 2017. In 2019, I ordered the Big Y-700.
The Big Y-700 is an entirely new test, not just an upgrade from the previous Big Y. YFull indicates multiple tests for the same kit on their tree. The new kit is displayed along with the old kit number.
When the Big Y-700 test was finished in 2019, Family Tree DNA removed the original Big Y test results and replaced them with the Big Y-700.
 |
| Two YFull kits for the same person |
When the Big Y-700 test was finished in 2019, Family Tree DNA removed the original Big Y test results and replaced them with the Big Y-700.
With the results from the first test gone, there is no way to compare the two.
But at YFull, the original test remains and can be compared to the linked Big Y-700 results.
I have also ordered a Full Genome test from Dante Labs. When those results come in, I can compare all three tests.
If we click Comparisons in the YFull menu, we can see the following Statistics tab that compares the original Big Y to the Big Y-700 test for this kit. The original Big Y was aligned to hg19 [this was before the Big Y-500 test], and the new results were aligned to hg38:
 |
| Big Y vs Big Y-700 |
The Novel SNPs tab below is especially fascinating. Position by position we see the hg19 and hg38 position numbers, all names that have been given to each SNP, and the calls from the two tests.
We can hover the cursor over any item to see an explanation.
For example, hovering the cursor over the green 1 shows that this is a Best quality SNP.
We want to examine one of these positions that was discovered in the Big Y-700 test but not in the original Big Y. Click on the yellow magnifier.
We can see that although this position was not discovered in the original Big Y, it was read 28 times in the Big Y-700 and has been given the SNP name FT85878.
We can also compare the STRs from the two tests.
 |
| Novel SNP comparison between Big Y test and Big Y-700 test |
We want to examine one of these positions that was discovered in the Big Y-700 test but not in the original Big Y. Click on the yellow magnifier.
 |
| New SNP discovered in Big Y-700 |
We can see that although this position was not discovered in the original Big Y, it was read 28 times in the Big Y-700 and has been given the SNP name FT85878.
 |
| More information about newly-discovered SNP |
We can also compare the STRs from the two tests.
The STR name is on the left, then the STR results from the first Big Y test for this kit, and finally the results for the same kit from the Big Y-700 test (the column on the far right).
Notice that on this first screen the STR results are the same for both tests:
However, as we move further down, we can see that they are not all the same:
In the above screen there were results for some markers in my original Big Y test [the second to last column], but no results for the same markers in the most recent Big Y-700 test.
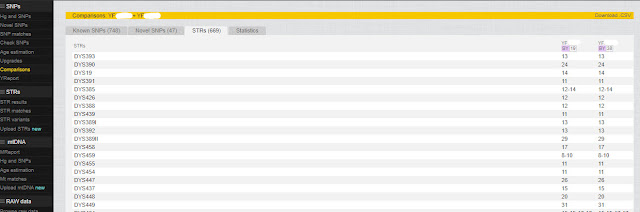 |
| Compare Big Y and Big Y-700 STRs |
However, as we move further down, we can see that they are not all the same:
 |
| Missing STRs in two versions of Big Y tests |
In the above screen there were results for some markers in my original Big Y test [the second to last column], but no results for the same markers in the most recent Big Y-700 test.
The same is true with the new test showing results for some markers that the older test did not.
Without being able to compare the two tests at YFull, I would not know about the missing results at any position.
But I what can I do about it? I may be able to order a test for a specific STR at YSEQ.
YSEQ can test a single STR or a panel of STRs. To order a single STR, click STRs in the menu on the left of the screen, then search to see if the STR is available for testing.
We can order a test for the DYS518 STR that was discovered in the original Big Y test, but lost in the Big Y-700:
Once you have received STR results from YSEQ, you can upload them to YFull and add them to the results you already uploaded. First click Upload STRs:
Now click UPLOAD STRs-YSEQ:
The ability to combine STRs from FTDNA and from YSEQ is wonderful.
Although we did not cover all of the benefits of YFull including SNP matches, STR matches outside of projects, estimating the dates when SNPs occurred, and many others, here are ten benefits that we did discuss:
1. You will not be charged for your upload until the results are ready, and you can add mtDNA Full Sequence results and multiple STR files for the same person at no additional charge.
YSEQ STRs
 |
| YSEQ STRs |
We can order a test for the DYS518 STR that was discovered in the original Big Y test, but lost in the Big Y-700:
 |
| Order YSEQ STR |
Once you have received STR results from YSEQ, you can upload them to YFull and add them to the results you already uploaded. First click Upload STRs:
 |
| Upload STRs to YFull |
Now click UPLOAD STRs-YSEQ:
 |
| Upload YSEQ STRs |
The ability to combine STRs from FTDNA and from YSEQ is wonderful.
SUMMARY: Why should I upload to YFull?
Here are 10 reasons
Although we did not cover all of the benefits of YFull including SNP matches, STR matches outside of projects, estimating the dates when SNPs occurred, and many others, here are ten benefits that we did discuss:
1. You will not be charged for your upload until the results are ready, and you can add mtDNA Full Sequence results and multiple STR files for the same person at no additional charge.
2. You can find additional information about your private variants including the names that have been given to this variant, the region of the Y-chromosome where it appears, whether it is available for Sanger testing at YSEQ, and more.
3. You can compare your results to people who tested at other companies.
4. You can compare your own results from different NGS or Full Genomes tests you've taken.
5. YFull Groups can display and compare all STRs (not just the first 111).
6. You can find SNP matches in YFull Groups, not for just your terminal SNP, but for any named or unnamed variant.
7. You can contact other people in your YFull Groups.
8. If you discover questionable SNPs or STRs in your NGS test, you can verify them at YSEQ and add them to your test results.
9. If some SNPs or STRs did not appear in your test results at all, you can order new SNPs or STRs from FTDNA or YSEQ and add them to your results.
10. Your results will not be wiped out, no matter how many versions of the same test you take.
You get a new, more comprehensive interpretation of your data. The benefits increase as YFull adds more features and more people submit results. Please seriously consider adding your results to YFull.
Update: See Advantages of submitting to YFull - Part 2
You get a new, more comprehensive interpretation of your data. The benefits increase as YFull adds more features and more people submit results. Please seriously consider adding your results to YFull.
Update: See Advantages of submitting to YFull - Part 2
What's next?
YFull is not just great for finding out more about your Y-DNA. You can also make great matches with mitochondrial DNA for your maternal line. See How to make matches with Mitochondrial DNA (mtDNA).