In my previous post Evaluating new Big Y changes I examined my updated Big Y results and the new chromosome browser. I could not examine the Big Y matching system because I had no matches. I do now, and I couldn't be more excited!
In this post we will see the entire process from examining STRs, ordering a SNP pack, and examining the Big Y. We will see how SNPs are determined, how to use the matching system, and how to use the new chromosome browser. We will also see what comes next.
STRs vs SNPs
When we first do Y-DNA testing, we usually get an STR test. We have traditionally considered only STRs to be genealogically relevant and have done SNP testing for deep ancestry.
For an explanation of the differences between STR and SNP testing see Y-DNA STRs, SNPs, and Haplogroups. This page will open in a new tab, so you won't lose your place here.
But are SNPs only for deep ancestry? Can SNP testing tell us anything about genealogical relationships that STRs can't?
But are SNPs only for deep ancestry? Can SNP testing tell us anything about genealogical relationships that STRs can't?
The Big Y test gives us both STRs and SNPs.
Let's start with examining 111 STRs and then see what information the Big Y added.
Let's start with examining 111 STRs and then see what information the Big Y added.
STRs led us to the immigrant ancestor
STR testing provided the clues I needed to trace my ancestor Electious Thompson back three generations further to his immigrant ancestor in Colonial Maryland.
To see how I did this, see Breaking through brick walls with Y-DNA.
After finding the common ancestor of my group within the Thompson project, I wanted to see if the STRs could place men within the known Thompson tree.
When you know that people are related, but you don't know precisely how, STRs can help sort people into distinct groups.
For example, in just the first 37 markers, there is a clear pattern emerging where descendants of Electious Thompson have a 20 at DYS458 and a 17 at DYS576.
The other men have a 19 and 18 at these positions. However, we often can't tell the order of the mutations.
 |
| Thompson DNA Project |
The current STRs can provide a broad, but not specific, grouping of descendants of our immigrant ancestor, Robert Thompson. More STRs could provide more detail.
However, the STRs could not lead us to the origins of Robert Thompson because we had no matches from any more distant European ancestors.
How can we find the origins of our immigrant ancestor?
If we've gone as far as we can with our genealogy, we need to find men who are more distantly related for Y-DNA testing. Many people wait for relatively-distant matches to show up in our surname projects, but this is not the best solution.
We may not find matches because many men may not realize the value of Y-DNA testing and therefore have not taken the test.
But even if they do take a Y-DNA test their results could have enough STR differences to cause these men to not show up on our list of matches.
Furthermore, they may have a different surname and may not join our surname project.
We may need to recruit people. We often need to look for people who do not share our surname because surnames were adopted throughout the world at different times and for different reasons.
Will we match people with different surnames who all came from the same small region?
How do we know which men to recruit for testing?
To find foreign origins, we often have to turn to SNP testing and haplogroup projects which reveal more distant relationships than STR testing and surname projects.
Men who join a haplogroup project have varying degrees of relationships, but they all share a distant common ancestor.
One Thompson took a SNP panel test and found that he belonged to haplogroup R-FGC11134. He joined the R-FGC11134 and Subclades haplogroup project.
Within the FGC11134 haplogroup project, we could see that the Thompsons could be, at least distantly, related to a man named Cairns. Cairns shared some distinct STRs with the Thompsons, but he did not show up on the STR matching list of any Thompson.
Cairns and Thompson STRs
Family Tree DNA has the following Expected Relationships chart in its Learning Center:
 |
| STR estimated relationships |
We can use this chart to help determine whether Cairns and Thompson are related. We will compare Cairns to my brother (Kit 38962).
37 markers
The results for the first 37 markers for Cairns and Thompson are as follows:
| 37-marker STR comparison |
There are STR mismatches at six locations, and two of these locations have a two-allele difference. The total genetic distance of Cairns and Thompson at 37 markers is eight.
To
be considered a match by Family Tree DNA, the genetic distance must be no
greater than four at 37 markers.
A genetic distance of eight is well beyond FTDNA’s match criteria of four, so these two are not a match. The Expected Relationships table confirms that they are “Not Related.”
67 markers
Cairns and Thompson have the following results for markers
38-67:
 |
| Comparison of markers 38-67 |
Family
Tree DNA considers a genetic distance of seven or less to be a match at 67
markers.
Thompson and Cairns have only one mismatch in the second
panel, bringing their total genetic distance to nine. Because the genetic distance is greater than
seven, neither of these men is on the match list of the other man. They are still
“Not Related.”
111 markers
Here are the results for markers 68-111:
| Comparison of markers 68-111 |
Cairns and Thompson have five more mismatches in the 68-111
panel bringing their total genetic distance to 14. This is beyond Family Tree
DNA’s cutoff of 10. Again, the Expected Relationships table states that they are
“Not Related.”
But Cairns and Thompson share some distinctive STRs. They could show up as SNP matches.
Cairns had already taken the Big Y and had been assigned to Haplogoup R-A9871, a subclade of R-FGC11134.
The Big Y is a SNP discovery test that finds previously unknown SNPs. SNP pack testing only looks for already known SNPs.
We know Thompson and Cairns share SNP FGC11134, but do they share more? Time to take the Big Y!
Cairns and Thompson SNPs
My brother's original Big Y results showed something far different from "Not Related." Cairns and Thompson shared 23 newly-discovered SNPs. This means they are definitely related.
When Big Y results are initially reported, the assigned haplogroup will be for a known SNP that is already on the haplotree. Family Tree DNA reported our shared haplogroup as R-A9871.
The novel common SNPs were not yet named; they were shown with their positions on the Y chromosome. So, instead of seeing a SNP name like R-A9871, we see a position name and the value that changed.
 |
| Haplogroup now listed as R-A9871 |
The novel common SNPs were not yet named; they were shown with their positions on the Y chromosome. So, instead of seeing a SNP name like R-A9871, we see a position name and the value that changed.
These results can be most easily seen in Alex Williamson's Big Tree. Cairns and Thompson are in the far right column. The first unnamed SNP shown below is 4780182-T-C.
 |
| Ales Williamson's Big Tree |
Naming the SNPs is not an automatic process, so it will take several days before the results are updated.
After Family Tree DNA manually reviewed the results, Cairns and Thompson formed a new haplogroup.
The 23 new SNPs were grouped together under the new haplogroup R-BY20951.
 |
| FTDNA haplotree |
The haplogroup appeared in our FGC11134 haplogroup project.
 |
| FGC11134 haplogroup project |
Even though we now know for sure that Thompson and Cairns are related, they can't be related too closely because the STR results are indicating that the relationship is distant.
There is no way to know the exact date of our most recent common ancestor, but the time can be estimated by SNP dating.
SNP dating is not an exact science, but it gets more precise as more tests are completed.
James Kane, administrator of the FGC11134 haplogroup project, did an evaluation of our results. His evaluations are important because he uses BAM files, has already been mapping results to Build 38, and has identified many SNPs that were not previously known. Kane estimated the date of our most recent common ancestor at about 1450 AD [TMRCA: 1450AD].
 |
| Estimated common ancestor date of 1450 AD |
Cairns has traced his ancestry to a specific parish in Scotland. Can we trace his line further? Will we have other testers in the future who will bring this date even closer?
Recruit another Thompson for Big Y testing
To help determine the accuracy of an estimated date for our common ancestor at 1450 AD, the next step was to test another Thompson for whom I had a known relationship. Thompson Kit 34484 (shown below) agreed to order the Big Y.
 |
| Haplogroup Project |
It is important for SNP dating to know the exact relationship between the two Thompson Big Y testers. Here is the Thompson line for kit 38962:
 |
| Family Tree for descendant of Electious Thompson |
Here is the Thompson line for Kit 34484:
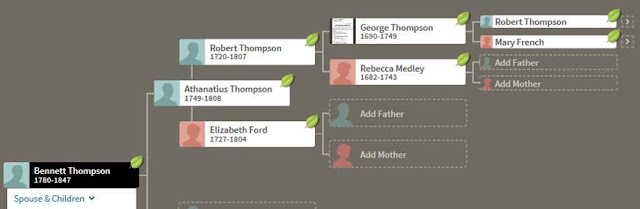 |
| Family Tree for descendant of Bennett Thompson |
Both men descend from George Thompson who was born about 1690 in Colonial Maryland.
Any SNPs that are shared between the two Thompsons, and not by Cairns, occurred sometime between the birth of George Thompson and the common Cairns-Thompson ancestor.
We can determine how closely related the Thompsons are to Cairns by seeing how many SNPs are shared between the two Thompsons. Any SNPs that are not shared can be assigned to the lines of James or Robert Thompson (sons of George).
The new Big Y results are in!!
We knew from STRs and genealogy that the Thompsons were related.
As expected, when the results of Kit 34484 first arrived, all three men (Cairns and two Thompsons) were assigned to haplogroup R-BY20951.
But I also expected that the Thompsons would share some SNPs that Cairns would not have. After the FTDNA manual review, the new haplogroup is now shown in the FGC11134 project.
 |
| New Thompson haplogroup |
The two Thompsons are now in haplogroup R-FGC65820, and Cairns is still in R-BY20951. But the review did much more than name a new haplogroup. The list of unnamed variants and the known SNPs changed on all three kits.
Unnamed variants
Here was my brother's list of unnamed variants before the FTDNA review. He had six unnamed variants:
 |
| Unnamed Variants before FTDNA review |
Here is his list of unnamed variants now. There are only two:
 |
| Unnamed Variants after FTDNA review |
What happened to the four that are missing?
They are now named and assigned to the haplotree.
Some of these SNPs had been named when I submitted my original Big Y test to Full Genomes Corporation. Three of these were found in the FTDNA review to be shared with Cairns.
Our haplogroup R-BY20951 now shows 26 SNPs:
 |
| FTDNA haplotree |
Position 11649109 is now SNP A18880; position 11321844 is SNP FGC65819, and position 19139783 is SNP FGC65831.
Only one unnamed variant was shared by the two Thompsons and not by Cairns. This was position 11514480 which is now called SNP FGC65820 and forms our new haplogroup.
When those four SNPs were named and placed on the haplotree, it left only two unnamed variants for my brother. These SNPs occurred somewhere in the line of James Thompson, son of George.
New Big Y matching system
We can examine other variants in the Big Y Matches section. When you go to your Big Y matches, you will see five haplogroup levels. Your list of matches at the lowest level will be shown along with all of your non-matching variants.
It is important to understand what constitutes a match. Family Tree DNA lists only those people who match within an average of 30 SNPs [this has also been stated as up to 40 total SNPs, 20 for you and 20 for your match].
We know that Cairns and Thompson share 26 SNPs that nobody else has, and they probably also have "private" SNPs (i.e. SNPs not yet seen in anybody else). So we will probably not see anybody other than Cairns and Thompson in this match list.
Please note the five haplogroup levels in the image below (FGC11134, ZZ44_1, A9871, BY20951, and FGC65820).
The two matches at level FGC11134 does not mean that only two people are in this haplogroup--there are hundreds. It means that only two people in this haplogroup are within 30 SNPs of my brother.
 |
| Big Y match list for haplogroup R-FGC65820 |
This screen shows only one match (the other Thompson) in haplogroup R-FGC65820. He and my brother have three non-matching variants. This tells me that the other Thompson has variant 8837670, and my brother does not have this. This variant occurred in the line of Robert Thompson, son of George.
The other two variants 12144610 and 56831461 are my brother's variants; they occurred in the line of James, son of George.
Where is Cairns? He is not within Haplogroup R-FGC65820, so he is shown at all of the higher levels. If I click on R-BY20951, I will see matches for both Thompson and Cairns.
 |
| Big Y match list for haplogroup R-BY20951 |
We see that Cairns and my brother mismatch on five named SNPs and four unnamed variants. So we may want to determine why these are not shared.
How are SNPs identified?
Now we get to the difficult part. How do we know if a SNP is valid? How do we know whether we have more or fewer SNPs than FTDNA identified?
We must first understand how SNPs are determined.
During the testing process, your DNA is not read in one continuous stretch. Instead, your DNA is broken into random fragments. The test reads these fragments from each end. Some fragments are read many more times than others.
After being read, the fragments are aligned to the reference sequence, and differences from the reference sequence are identified. The difference from the reference value is your "derived" value.
Unfortunately, not all of the reads may give the same result. One read may show the reference allele (for example a C) and another read may show a derived value (for example a G).
When fragments were aligned to the old Build 37 reference sequence there were more varying calls reported, often due to poor alignment. There will be more valid SNPs identified now that Family Tree DNA is using the more recent and more accurate Build 38.
To be considered a high quality SNP by Family Tree DNA the position must be read at least ten times. The number of differing calls is then taken into account.
A position that was read a few times with different results will be considered to be a much less reliable SNP than one that was read many times with a consistent result.
How can I tell if my SNPs are reliable?
The best, and easiest, way to analyze your results is to send your file to third-party analysts like YFull or Full Genomes Corporation, or both! There are also many haplogroup administrators and other volunteer analysts who will do this for free.
I recommend using as many analysts as possible because all will contribute to your understanding.
FTDNA has a file called the BAM file which contains the information needed to do a full evaluation. I will have a future blog post showing results from BAM analysis.
But Family Tree DNA provides a tool that will help you to do some analysis by yourself. When you are trying to find out if you have a particular SNP, or why a known SNP is on your list of non-matching variants, you can use the chromosome browser. You can see in the browser how many times a position was read and if all calls were consistent.
Below we will see examples of three different types of reads: one position that was read fewer than ten times, another position that was read more than ten times but with different calls, and a third position where there are two SNPs.
Evaluating variants with the Chromosome Browser
When you go to your Big Y results page, you will see three tabs for Named Variants, Unnamed Variants, and Matching.
 |
| Big Y Named Variants tab |
Under the Named Variants tab, you will see the Y-Chromosome Browsing Tool (I will hereafter call it the Chromosome Browser).
Below that you will see columns for SNP Name, Derived Status (whether or not you have that SNP), whether the SNP appears on FTDNA's Y haplotree, the hg38 Reference value, your value (which is called Genotype) -- it will be different from the reference value if you are derived for that SNP, and Confidence (how confident FTDNA is that this is a valid SNP).
 |
| Y-Chromosome Browsing Tool |
A SNP with low confidence
In the SNP name search, I first entered my haplogroup FGC11134. I saw "Currently no results."
 |
| SNP Name search box: Currently no results |
That didn't make any sense. I know my brother should have that SNP.
I noticed that the default for the "Derived?" column is set to "Yes." Since there were no results for that SNP, FTDNA was telling me that my brother had the ancestral value, not the derived value.
So I reset the Derived column to "Show All" and searched again for FGC11134.
This time FGC11134 showed up with a question mark in the Derived column. The Reference is T and the Genotype [Derived value] is "?".
 |
| Questionable SNP |
I can click on the name of the SNP and view it in the Chromosome Browser.
 |
| Click link to view SNP in Chromosome Browser |
Clicking on the SNP name takes you to that position in the Chromosome Browser.
The Chromosome Browser shows that SNP FGC11134 is position 19221580. We can see the Reference value T highlighted in red below the black arrow. The four calls below it (in pink) are all A. This means that this position was read four times in my brother's DNA test. My brother had "A", and not the Reference value which is T.
 |
| Chromosome Brower results |
But because this position was only read four times, it does not qualify as a reliable SNP call.
To be considered a valid SNP by FTDNA, the position must have been read at least ten times. So instead of the reported value of A, the derived value is listed in my brother's report as "?"
Most third-party analysts would report this as a SNP.
For example, YFull will report variants that have been read as few as two times. It will rate these variants based on their reliability, but will only name them as SNPs and place them on their tree when shared with others. YFull would have been reported FGC11134 as a SNP on my brother's report.
Although FTDNA did not report this SNP because it did not show up with high enough confidence, it does appear in the Chromosome Browser, and my brother is still placed in the FTDNA haplotree within FGC11134.
A SNP with different calls
We can examine the non-matching SNPs between Cairns and Thompson using the same process.
For any named variants (BY26955, FGC22107, etc.) we use the Named Variants tab and the SNP Name search.
For unnamed variants (7403088, 12144610, etc.) we can only view our own variants using the Unnamed Variants tab and then clicking the Position name.
I want to see why BY26955 shows up as a Non-Matching Variant between Cairns and Thompson.
In the Named Variants tab, enter the SNP name into the SNP Search box. Notice that the Derived column contains "Yes", so Thompson has this SNP, but Cairns must not.
Now click the name of the SNP (the blue link).
 |
| Search for SNP BY26955 |
The Chromosome Browser shows that SNP BY26955 is position 10658444. Here the reference value is C, and the derived values below it are mixed.
 |
| SNP BY26955 in Chromosome Browser |
This position appears to have been read 14 times. Some of the lines are in vivid colors indicating High Quality. Some are faint indicating Low Quality.
Eleven calls are A, two are T, and one is the Reference C (where the cursor is pointing). But the Genotype is reported as A with High Confidence.
Did Cairns have even more mixed calls at his location causing the SNP to be considered low-quality in his results?
A position with two SNPs
SNP FGC46559 appears on the Cairns and Thompson list of Non-Matching Variants. I searched for this SNP in my brother's account:
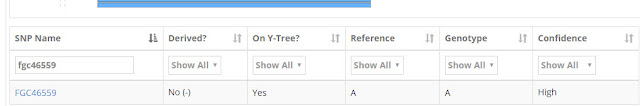 |
| Search for SNP FGC36559 |
As you see above, Thompson is not derived for SNP FGC46559. The reference and genotype are both listed as A which means that Thompson has the ancestral value and does not have a SNP here. Since it is a non-matching variant, Cairns must have this SNP.
This one is puzzling. When we click the SNP name and go to the Chromosome Browser, we see the following.
 |
| SNP FGC46559 in Chromosome Browser |
There are many more calls for this SNP that are not shown. You have to scroll up and down to see them all. This position was read a total of 76 times.
The Reference value is A and is highlighted in red directly below the black arrow. All of the Genotype calls beneath it (in pink) are G except for that one blank space on the seventh line from the top. Yet the Genotype and Reference are both stated to be A.
Vladimir Semargl of YFull answered this question. SNP FGC46559 is the mutation 19714591 A>C. But this kit has 19714591 A>G which is named FGC65832.
So my brother is negative for FGC46559, but positive for FGC65832. We can see these two SNP names at YBrowse:
 |
| YBrowse |
For an update on this SNP, see Great updates to YFull.
How many more SNPs need to be reviewed?
The best results will be obtained when we get the BAM file and submit it to multiple third party analysts.
I have had my brother's previous Build 37 results analyzed by several third parties.
My personal favorite is YFull because the results are online and can be compared to others. YFull finds many SNPs not reported by FTDNA, reports a few hundred STRs that can be extracted from the Big Y test results, places you on their haplotree where you can see close as well as distant matches, and more.
You can see what their evaluations look like in What are the benefits of YFull? and Big Changes to YFull.
What did we accomplish with Big Y results?
Only the Big Y could give us the following results that cannot be obtained by STR testing or SNP pack testing.
- Proving relationships: STR results showed that the Thompson kits 34484 and 38962 are probably related. SNP results prove they are definitely related. Knowing that they have at least three unshared SNPs helps to verify the known genealogy and show why they may not show up as Family Finder matches. With the Big Y we found that although STR matching indicated that the Thompsons were not related to Cairns (at least not within 15 generations), they are definitely related to Cairns, and possibly within that time period.
- Moving SNPs forward in time: Before the Big Y we could only order individual SNPs or a SNP pack. SNP pack results placed the Thompsons at haplogroup FGC11134 which James Kane dates at approximately 1350 BC. Big Y testing discovered 26 previously unknown SNPs that are shared by Cairns and Thompson. This moved the estimated date of our common ancestor from 1350 BC to 1450 AD!
- Naming new haplogroup subclades: To be related, men have to share the same ancient haplogroup. Before the Big Y, all haplogroups were ancient, so Cairns and the two Thompsons shared haplogroup FGC11134. But with the Big Y, haplogroups are moving into more modern times. Cairns and Thompson are still related, but because of the new Thompson Big Y results Cairns and the two Thompsons now have different haplogroups: FGC65820 is a subclade (subgroup) of BY20951.
- Contributing to science: We contributed to science by identifying two new haplogroups (BY20951 and FGC65820) and placing 27 new SNPs on the human haplotree.
- Dating a modern SNP: We identified one SNP shared between the Thompsons and not with Cairns. SNP FGC65820 ocurred no later than 1690 AD (the latest possible date of birth of George Thompson) and no earlier than the time of the most recent Cairns-Thompson ancestor (1450 AD?). Haplogroup FGC65820 is not an ancient haplogroup; it is within the genealogical time period.
- Separating descendant lines: We identified three Thompson Y-chromosome positions that mutated after 1690 and know in which lines they occurred. There will probably be more of these when the BAM files are analyzed.
- Narrowing ancestral origins: We know that Cairns came from a specific parish in Scotland, and although it currently appears unlikely, it still may be possible to find the common Thompson-Cairns ancestor. At the very least we may be able to narrow down the place of origin for Robert Thompson.
What can we do to further enhance our Big Y results?
- Recruit more testers within our surname groups. To leave a lasting Thompson legacy, I will be trying to recruit more Thompsons to take the Big Y test so that we can identify SNPs that have occurred since our common ancestor. Each new SNP match will create a new Thompson-specific haplogroup. We also have Thompsons who cannot trace their ancestors back to Robert Thompson but know by STR testing that they fit in somewhere. By taking the Big Y they may be able to find precisely where they belong. Cairns has already had another family member tested and is awaiting the Big Y results, so he will soon have a Cairns-specific haplogroup.
- Submit our results to third-party analysts. Evaluation of the BAM files will identify new SNPs and STRs. Further testing of other Thompsons, for example, can allow us to place specific SNPs and STRs with specific ancestors.
- Recruit people outside our surname groups. Look at STR matches to men with other surnames. In contrast to what many used to think, all of these different-surname matches are not caused by "non-paternal events." Some are caused by convergence where STRs mutate in one generation and back to the original value in another. STR convergence makes men look much more closely related than they really are, but SNPs can show the true relationship. Look at people in our haplogroup and geographical projects. Participate in the Activity Feed for these projects. By focusing on these non-surname matches and encouraging them to test the Big Y, sometime in the near future some men will share some SNPs with Cairns and Thompson, but not all. This will break up that large 26-SNP block of shared SNPs and help date them. Maybe one man will have a well-documented genealogy and share SNP FGC65820 (the one that occurred sometime between the common Cairns-Thompson ancestor and the year 1690). I'm sending my vibes out into the universe to find that man!
- Submit our new SNPs to YSeq to be made available for Sanger testing. We can verify questionable SNPs and also make it far less expensive for other men to find out if they are related.
What will happen next, even if we don't do anything?
- Big Y results will be much more meaningful next year. One reason is because many more people are now testing the Big Y, and so we will see greater refinement of the human haplotree. As Big Y testing becomes more mainstream many haplogroups will move into the genealogical time period. Another reason for better results is that all analysts will be mapping to hg38. Still another reason is that more tools are continuously being developed by FTDNA and by third-party analysts.
- Family Tree DNA has announced that they will be reporting STR results with the Big Y results sometime next year. With the ability to match SNPs and hundreds of STRs we will be able to make better genealogical conclusions.
There are other companies that offer a more comprehensive test of the Y-chromosome, but it is the potential to make matches that makes all the difference.
You can find existing matches with the Big Y matching system, and you can find and recruit others for future Big Y testing by using Family Tree DNA's STR matching system and by joining a wide variety of FTDNA projects.
We're heading to a great future. The Big Y can prove modern genealogies and give us our ancient ancestry as well.
We're heading to a great future. The Big Y can prove modern genealogies and give us our ancient ancestry as well.
Until whole genome matching is widely utilized, the Big Y could be the best DNA test ever!
Next: See another case study in which I was able to find the exact European origin of my ancestor in The Amazing Power of Y-DNA.
Next: See another case study in which I was able to find the exact European origin of my ancestor in The Amazing Power of Y-DNA.

16 comments:
Wonderful article, Linda. Thank you very much for taking the time to give us this clear and thorough example. Propagating for others to find: https://www.wikitree.com/g2g/519812/new-blog-post-by-linda-jonas-describes-ftdnas-big-test-detail.
Thank you for this Article. It is a very good read for the new Big Y conversion. I am sharing the link far and wide!
I agree this is a well written article and I have learnt how to examine my results with the new Browser. I'm in a very small group under Z253>L1066>CTS9881>BY3930, I have my third cousin tested to SNP BT3930 and would have bought the Big Y this time had FTDNA had not applied the restriction for managed kits. I contacted my third cousin to explain the new rules and extract a written permission to proceed, but he has declined to engage with me for some reason. We know who our common ancestor is so that's at least one good thing has been determined. Our other match has Big Y, but we are 63/67 STR markers ans have a number of non shared SNPs, I tried to extract the remaining FTDNA STR 68-11 from our Y full results but not all have reliable reads so can not be precise on what our differences are at 111 probably in the order of 9 differences.It would be great to be able to investigate more closely where our match and our line split, It is encouraging that we all share the same surname and originate from Ireland. I hope that the new year will bring better fortune and possibly another to compare with as I think two Big Y results are not enough to establish more, is that correct?
Mike, You cannot extract the 68-111 panel of SNPs from the Big Y results. The Big Y does not read all of them. I looked up your haplogroup on the Big Tree, and you do have a fairly large number of non-matching Barry SNPs, so it would be great to find another Barry for Big Y testing. To find more Barrys, be sure you have an online family tree, and trace descendants of your lines so that others can connect their ancestors to yours. Post screen shots of your DNA results to the profile pages of your Barry ancestors. For example, you might post a screen shot of the CTS9881 page from the Big Tree and explain what it means. These images encourage other people with your surname to contact you. I get questions like, "I don't see anything like this in my Ancestry DNA results. How did you get this?" Then I tell them about Y-DNA testing. Even though you already submitted your Build 37 results, be sure to submit your new Build 38 results to the Y-DNA Data Warehouse at http://www.haplogroup-r.org/submit_data.php
If you can afford it, you might also consider contacting your Big Y match and offering to contribute to, or pay for, the upgrade of his results to 111 markers.
Contact me by email if you have any questions.
This is my favorite blog post of late. Fantastic and thank you, Linda! I'm also sending out positive vibes that you find your man with the well-documented genealogy who shares SNP FGC65820. :)
I've had a general understanding of the importance of the Big Y test, but this has really been enlightening for me, helping me pull the pieces and site functionality together.
I am curious to learn more about the efforts to date SNPs (that you were able to do that back to 1450 is remarkable), and would welcome your input on that front.
The SNP dating was not done by me. Dating can only be done with a substantial database of kits to compare. This particular SNP dating was done by James Kane who has a very large database at http://haplogroup-r.org/
James Kane can probably date Haplogroup R SNPs as well or better than anyone.
I really appreciate the combined vibes. They will get to that man faster than mine alone!! I laughed pretty hard when I saw that.
Thank you.
Nice article! Thanks for posting! In the section "Why is FGC46559 not reported?" you discuss why the results are puzzling and say in cases like this, we should submit this to FTDNA for further evaluation. What is the form or process we use to submit these puzzling calls to FTDNA?
Thanks.
If you want to have something in your own kit evaluated, contact Michael Sagar at Family Tree DNA and give him you kit number. You use the same procedure if you want two kits compared. Michael Sagar is very responsive, and usually will have it done within a day.
Thanks for your clear explanation, I learned a lot. However, I still have some doubts, as I got my BigY results yesterday:
My haplotree says “your confirmed haplogroup is: E-CTS1096” (which hasn’t changed since I imported my results from Geno 2.0). However, down the tree it says E-S11956 (marked in green), and below that, it still offers me individual SNP tests.
If I look up CTS1096 on the SNP search box with all options on "Show all", I get this:
CTS1096 ? Yes T ? High
and "No reads found" if I click on the blue SNP name.
How can that be? how could I belong to that haplogroup I that SNP wasn't read?
On the other hand, S11956 gives:
S11956 Yes (+) Yes C G High
So, there seems to be a mismatch between my haplogroup and my terminal SNP, even though from what I have seen here and other places they should match. Am I wrong or missing something?
Thanks a lot for looking into this if you have the time, I would appreciate any input you may have.
jlperello: There are many reasons for this, and you may discover some of it by investigating your individual SNPs. But one of the biggest reasons, as I have stated, is that haplogroups do not automatically change with the Big Y testing. Also, there are known glitches in the new system that Family Tree DNA has discovered and is in the process of fixing.
You could appear in a haplogroup where the SNP was not read if you tested positive for known downstream SNPs.
If you have any matches, you can contact them and compare unnamed variants. That's how Cairns and I discovered that we shared some. After that, FTDNA looked at the kits, named the SNPs, and updated the haplogroups.
Thanks Linda for your reply.
Unfortunately I have no matches at all, so it seems the only thing I can do for now is to wait.
Also, on Yfull E-CTS1096a and E-S11956 are on the same spot, so it doesn't look as if I have tested positive downwards.
Well, patience.
Maybe the BAM and Ytree can shed some light on this.
Thank you.
Thanks for posting. Some interesting information in the article. Points to share about the STR markers chart listed from FTDNA is that it should be used as a guidepost vice a fact and that a Y67 marker test appears best to use as a minimum to determine potential close relationships. As an example, testing at Y111 markers, several surname group members matched me at a 0-1 genetic difference. According to the FTDNA chart, those surname members should be a close relation to me. When BigY testing was complete and our SNP was compared, some members at a close genetic distance to me, did not belong to my genetic subgroup within the surname group. That was a surprise, but it shows that STR testing alone is not enough to properly shape a surname group's family tree.
I am very much a novice though I have done extensive DNA test.Would appreciate any comments and guidance regarding recent testing.
At FTDNA I have 2 35/37 matches with the same surname (Parr) but one that is different from mine(Bartlett). Neither have done testing beyond the 37 marker Y-DNA test at FTDNA. I had my Big Y results evaluated at YFull. My terminal SNP was determined to be S2077. Both Parrs agreed to test my terminal SNP at YSEQ which was S2077. Both were a match for me.
I next took my novel SNPs identified by YFull, of which there were 10, and checked with YSEQ to see which could be tested there. It turned out that 6 of the 10 could be tested.
I agreed to pay for the tests. Unfortunately only one Parr responded so I went ahead with the tests. These were the results:
BY13501 G+
BY13535 A+
YFS252509 C+
YFS252553 C-
YFS252558 C+
YFS252559 T+
So we matched on 5 of the 6.
So the questions I now have are the following:
1. Are these results only genetically interesting?
2. Do they provide substantial proof that we, the Parrs and Bartletts, are definitely related within a reasonable time frame that has some chance of being discovered?
The reason I've gone to the extent that I have with testing their DNA is no one has been able to trace our line back to Europe. Our "brick wall" is a Bartlett born in 1743 in Massachusetts for which a solid paper and DNA trail exists.
The Parrs are of interest because both have good paper trails to England and Ireland. Additionally they have no DNA matches with any of the other Parr lines that have tested.
The other interesting fact is that we share certain STR values
That are "odd" for our I1a Z63 Haplogroup.
Any suggestions would be gratefully received.
Great blog... This blog share valuable information on DNA testing and its importance. Genetic testing also help to detect future health conditions. Thanks for sharing valuable content.
Very informative blog post. This blog share complete information on genetic testing. Best genetic testing is a type of medical test that identifies changes in chromosomes, genes, or proteins. Thanks
Very informative and well written post! Quite interesting and nice topic chosen for the post Nice Post keep it up.Excellent post.I want to thank you for this informative post. I really appreciate sharing this great post. Keep up your work. You did a wonderful thing and should be proud of yourself for sharing. Thank you! I am very happy to your post its a great post,I'm glad you enjoyed the post.Keep sharing such useful informations. So informative blog, thanks for providing valuable information
Post a Comment