I have received my brother's Big Y results from Family Tree DNA. You can see the incredible discoveries I made by seeing my post The Big Y test from Family Tree DNA can knock your socks off! My brother, who is a Thompson, shared several novel SNPs with a man named Cairns, and we formed a new haplogroup. One of my distantly-related Thompson relatives then ordered the Big Y, and we are awaiting the results. Can I find out more that might help us by submitting my results to YFull.com?
Getting Started
If you took the Big Y from Family Tree DNA, you can submit your BAM file to YFull. Your BAM file is not automatically generated by FTDNA; you must request it first by going to your Big Y results section and clicking Download Raw Data.
Go to the bottom of the next screen to request the BAM. You will receive an email when your file is generated.
Once your BAM file is ready, go back to the screen above and again click Download Raw Data. On the next screen scroll to the bottom where you will find these links:
Do not click Download BAM because the file is too large. Click Share BAM, and you will receive a temporary link that you will share with YFull.
Now go to YFull.com to order the interpretation of your test results. Your first thought might be, "I just spent a lot of money on the Big Y test. I don't have another $50 to spare right now." Here's the good news. You don't pay up front. They will send you an invoice when the interpretation is finished. So you can order now, pay later.
Our results are in!
When your YFull results are returned, this will be your home screen. There are three main sections: Haplogroups and SNPs, STR results, and Novel SNP results. On the left of the screen are links to even more tools.
There are so many resources that we will cover only a few of them. Otherwise this blog post would take you hours to read.
Haplogroups
First we will see where this kit fits into the human Y tree. Go to the SNP menu on the left of your screen and click Y Report. This will show your haplogroup at the top [here R-A9871] then show your decent from Y-chromosome "Adam". The top of the report is shown in the next image.
Scroll all the way down to the bottom to see your most recent placement on the tree:
On the haplotree at Family Tree DNA, my current haplogroup is R-BY20951 which is the new haplogroup formed when my results were compared to those of my Cairns match. It is further down from R-A9871 and more recent in time. But YFull is only reporting results for kits that are in their system, and Cairns is not yet in the system. So I am still at R-A9871 here.
SNPs
Because the Cairns results are not in YFull this means all the SNPs I shared with him are considered by YFull to be novel ("private") SNPs seen only in my results. We will click on Novel SNPs to see the reports.
YFull's reporting of SNPs is quite a bit more extensive than FTDNA's. FTDNA reported a total of 26 SNPs in my Big Y results. 23 were shared with Cairns, and three were considered to be novel or "private" variants seen only in my results. YFull found a total of 48 SNPs which were categorized as best quality, acceptable quality, ambiguous quality, and low quality. The best-quality SNPs are shown in the next image.
In the image above the SNP locations are shown as well as the names they have been given. Both Full Genomes Corp and FTDNA had previously named many of these new SNPs. This is one of the problems with SNP naming. There is no consistent naming pattern, and SNPs are sometimes named multiple times. The SNPs starting with the letters BY were named by Family Tree DNA. The ones starting with the letters FGC were named by Full Genomes Corp. There are 12 Best quality SNPs. Two of these were not reported by FTDNA.
Here are the 19 acceptable quality SNPs. Some were not reported by FTDNA.
There are 15 ambiguous SNPs. Only one was named by FTDNA.
There are also two low quality SNPs. Family Tree DNA did not report these.
You can find details about each SNP and why it was categorized as best, acceptable, etc. by clicking on the other tools. The important issue is this: Family Tree DNA is conservative when identifying SNPs. YFull is reporting several more, but some of these may not be reliable SNPs that can be branch markers on the Y-haplotree. However, when any of these are shared with someone else they can be. Some of these newly-identified SNPs may match Cairns. Others will probably match my Thompson relative. We will no doubt discover new SNPs and be able to determine our relationship more closely when those two additional kits appear on YFull. We can verify these SNPs by testing them at a company called YSEQ.
YFull is also attempting to date all SNPs on the haplotree. This dating is based on the kits that have been submitted to the tree and will become more accurate as more test results are submitted.
STRs
Now, another big discovery. I have tested 111 STRs at Family Tree DNA which is the maximum I can order. New STRs are discovered in your Big Y test, but they are not reported by Family Tree DNA.
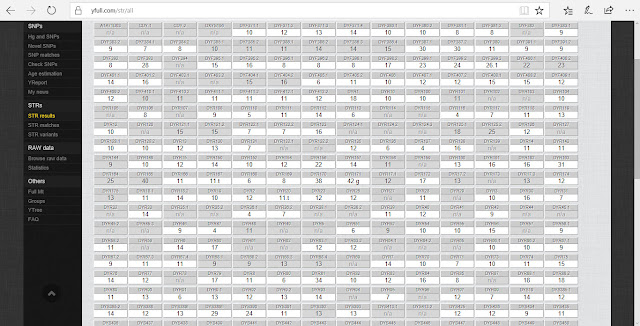
YFull can report approximately 500 SNPs from your Big Y test! Here is a portion of my STR results:
These STRs are compared to the results of other men and I can see a list of matches. I don't have any close matches yet, but I have many distant matches:
If any of these look interesting I can contact the kit owner by clicking the PM (Private Message) link. The link is shown as a blue envelope next to the "Most distant ancestor" column. YFull is serious when they call it a Private Message. If I click the envelope link I see a screen like this
I can hardly wait until I compare my new STR results to my Thompson relative!
Summary
We have touched on only a few of the many benefits of submitting your results to YFull. The system is very user-friendly. Even though it's easy to use, you can get as technical as you want by using the various tools. Their analysis tools are quite extensive and well beyond what I have seen elsewhere.
You will discover new SNPs and new STRs. You may also discover new matches because YFull accepts results from Family Tree DNA as well as Full Genomes Corp.
You will also be helping to build the Y Tree and to more accurately date the various branches.
As with all DNA testing for family history, our results are meaningful only when compared to others. I strongly encourage all of you to submit to YFull.
Update: See Big Changes to YFull
2019 Update: See Advantages of Submitting to YFull











7 comments:
Can you see any point testing my 4th cousin Anderson.
We match at Ftdna y111 GD4 and aitosomally
I have uploaded to Yfull and am in the BigTree in the scots cluster kit anderson 16273
Cheers
Mike Anderson
Whether or not you order a Big Y test for your fourth cousin depends on what you want to accomplish. A match to your fourth cousin will help you to define where SNPS occurred in your Anderson line and will help you to determine the validity of some of the STRs reported by YFull. When you know which STRs and SNPS were held by your ancestors you can more closely determine how other people are related to you.
On the other hand, if you are mainly interested in extending the line of your ancestor William Anderson (born ca.1660), then your efforts may be better spent recruiting more distantly related Andersons to do Y-DNA testing.
Read my blog post "The Big Y test from Family Tree DNA can knock your socks off!" at to see my testing strategy. After the Big Y results are completed for my most
distantly-related Thompson, I will next attempt to test a fourth cousin so that I can do what I said at the beginning: see in which generation SNPs occurred in my Thompson line and determine the validity of STRs reported by YFull. When I have a distantly-related Thompson and a more closely-related Thompson to compare results to, I will have a much better picture of the DNA profile of my ancestors.
I have two issues with Yfull's 'STR matches' and the age estimation methodology:
1) My 500 listed STR matches have no relevance to my sample because they are ALL outside my haplogroup (R-Z37492), and therefore of greater genetic distance than those within it. Five samples are within this haplogroup, but none make it to the '500 listing' ie all calculated at greater than Yfull's genetic distance cut off = 0.24. How is it possible for these more closely related samples to be at greater genetic distance than samples outside our haplogroup R-Z37492? I therefore do not believe their age estimation methodology, especially after considering issue (2). I have pointed this out on Yfull's facebook forum but received no response.
2) I submitted 2 samples to Yfull, one from the FTDNA Big Y test and another from FGC Y-Elite 2.1 test ie from the same person; they calculate a TMRCA between these two samples of 325 yrs!! I could understand an error bar of +/- 50 yrs (1 mutation) or, at most, 150 yrs but their age estimates seem x2 what they should be because they consider genetic distance to reflect a single branch on the tree (as for SNPs) and not 2 branches for any two samples (STR Infinite Allele method) - this may be the reason none of the same-haplogroup samples mentioned in (1) made it to my 'top 500 listing'.
I’m interested in YFull’s service primarily for the SNP dating analysis, but wonder about the comments raised by DjH above. I recognize this is a burgeoning science and there’s still much to learn and work through (and that more testers help that effort). Are the age estimates/timelines for SNPs updated as new testers or approaches are developed (hopefully addressing point like that raised by DjH) or are the estimates static?
SNP dating is adjusted as more samples are returned.
Looking at R-BY20951 on yfull today, I notice there's a kit there #YF15578, which is from the Ukraine. That seems very far from all the other kits under R-FGC11134 which seem to be mostly from Ireland and Britain. Do you happen to know anything about that? Very intriguing, no?
Patrick, yes, we have been in contact with him. We have another test on the way.
Post a Comment