Nebula Genomics offers "Deep Ancestry" as part of its Whole Genome Sequencing test. Your results for Y-DNA and mitochondrial DNA (mtDNA) can be transferred to YFull to get these ancestry reports.
Everybody has mitochondria. So in this article we will discuss mitochondrial DNA.
We will examine what mtDNA is and how it is inherited. Then we will find out how your mtDNA is interpreted and what you can learn about yourself and your ancestors.
I am going to tell you everything you need to know. Nobody has ever gone into this much detail about mitochondrial DNA at YFull, so get ready!
You'll even see that this is more than about deep ancestry. You will also be given tools to find the names of modern ancestors!
What is mitochondrial DNA and how is it inherited?
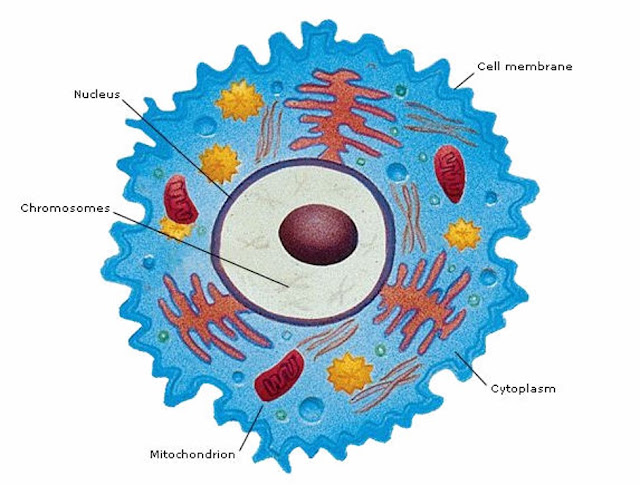
Your 23 pairs of chromosomes are located in the nucleus of the cell. Most genetic testing focuses on this nuclear DNA.
Mitochondria are organelles found outside the nucleus of the cell. They carry their own DNA.
Mitochondria produce energy for cells. You have more mitochondria in cells that require more energy.
Mitochondria are inherited from your mother [however, there are known cases where someone has inherited mitochondria from both parents].
Both males and females inherit mitochondrial DNA from their mother, but only females pass the mtDNA to their children.
So your mitochondrial DNA can be used to find out more about your strictly maternal line: your mother's mother's mother . . . .
 |
| mtDNA inheritance is shown in red Below is a four-generation photo of a great-grandmother, grandmother, mother and her seven children. All of these people inherited the same mitochondrial DNA: |
The seven great-grandchildren are now grown and have children of their own. The boys in the photo did not pass their mitochondrial DNA to their children. The girls did.
However, only one of the four great-granddaughters has a daughter. So only one living descendant can pass this mitochondrial DNA to her children.
This example shows why it is important to preserve your mtDNA for future generations.
What is YFull?
YFull is not a DNA testing company. YFull is an analysis and comparison service for Y-DNA Next Generation Sequencing and full mitochondrial DNA sequences.
These interpretation and comparison services are very different from those found at any DNA testing company.
Why would you want to submit your
mitochondrial DNA results to YFull?
At YFull, your results will be compared to scientific samples and to people who tested at different companies.
YFull only began to do mitochondrial DNA analysis in 2019, and so many people do not know about its mtDNA services. It is possible that you may not yet have many matches there.
However, even though many of YFull's mtDNA services are just starting you will learn a lot about your mitochondrial DNA from YFull's matching system and YFull groups.
How to transfer your mtDNA results to YFull
I showed how to transfer mtDNA results from Family Tree DNA here: How to make matches with Mitochondrial DNA (mtDNA).
If you took a full genome test from Nebula Genomics the full mitochondrial DNA sequence can be extracted from the test and automatically transferred to YFull.
Nebula Genomics describes the following benefits of YFull:
- Find matches based on mtDNA
- mtDNA haplogroup determination
- Identification of all mitochondrial SNPs
- Positioning in MTree
- Regular updates
To submit your mtDNA results from Nebula Genomics, see the Deep Ancestry section:
The amazing thing is that if you submit through this screen, you will obtain all of the YFull mitochondrial DNA benefits absolutely free!
MITOCHONDRIAL DNA RESULTS AT YFULL
Determining your results
Over time, mutations have occurred in your mitochondrial DNA.
When your mitochondrial DNA is tested, your results will be compared to a reference sequence. Your report contains the different mutations between you and the reference sequence.
For many years, mtDNA results have been compared to the Cambridge Reference Sequence. This was the first full mitochondrial DNA sequence, and it was completed in 1981. The Cambridge Reference Sequence is referred to as the CRS. A revised version is the rCRS.
Several years later a sequence was compiled to represent "Mitochondrial Eve." This sequence is called the Reconstructed Sapiens Reference Sequence and abbreviated as RSRS.
You will usually see two sets of mitochondrial DNA results: one compared to the rCRS, and one compared to the RSRS.
The mitochondrial DNA tree
Your results will then be compared to other results, and you will be assigned to a haplogroup which is a group of people with similar results.
Your results will be placed on the mitochondrial DNA tree which starts at "Mitochondrial Eve" and branches down to the present day. You will see exactly where you fit in.
"Mitochondrial Eve" was not the first woman who ever lived, nor was she the only woman alive at the time, but she is the only one who has an unbroken line of descendants.
YFull has a tree called the MTree where you can see the current mitochondrial DNA tree with its branching haplogroups.
The very top of this massive tree is shown below. "Mitochondrial Eve" is in haplogroup L. Neanderthals are listed before "Mitochondrial Eve."
You can see your mtDNA results, learn how you fit into the great mtDNA tree, and even see matches by transferring your results to YFull.
Your mitochondrial DNA results
When you submit results to YFull you will see a menu of options for Y-DNA and for mtDNA. Here are the current options in the mtDNA menu:
 |
| MtDNA menu |
OK, right away it looks complicated. Don't worry, we'll examine each of these and see what you can discover.
Haplogroup and SNPs
The first section is Hg [abbreviation for haplogroup] and SNPs [abbreviation for Single Nucleotide Polymorphism]. What do these terms mean?
Hg is an abbreviation for haplogroup. A haplogroup is a group of people with similar results. Mitochondrial DNA results have shown that all people alive today descend from "Mitochondrial Eve." The various haplogroups show the trail from you back to her.
Your results placed you in a haplogroup which is then put on the mitochondrial DNA tree (the MTree shown above.)
A SNP is a type of mutation. A Single Nucleotide Polymorphism is scientific name for a mutation where one value mutates to another.
For example, you may see a mutation like A769G. This means that at mitochondrial position 769 the reference sequence had an A, and you have a G.
In addition to SNPs you may have other types of mutations such as insertions and deletions (called indels).
An indel is not a change from one value to another. It is deleting that position altogether or adding additional ones.
We will see a SNP and an indel in the example below.
Haplogroup and SNP results
When you click on Hg and SNPs you will see a screen similar to the following. Only the first three lines are shown below.
Your mutations
 |
| Haplogroup and SNPs |
Your mutations
Here is a closeup of the SNP and indel information on the right of the screen:
These mutations show how your results differ from the Reference Sequence. They show your mutation and its location in the mitochondria.
For the first mutation on the list, the Reference Sequence has A at position 302. This is called the Ancestral value (Anc). If these were your results you have ACC which is called the Derived value (Der).
Your mtDNA has two additional mutations (CC) at position 302. These are insertions in your mitochondria, so this is an indel.
In the next row, the Reference Sequence has A at position 15052. Your derived value is G. So the mutation is listed as A15052G. This is a SNP.
Notice at the top of the image that you can download three versions of your results at any time.
You can see several visual representations of any mutation by clicking on the FASTA link next to the mutation.
Here is an example. In the screen below, a mutation from C to T is highlighted. There is another mutation at two positions further.
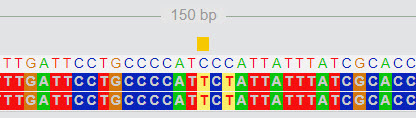 |
| FASTA Viewer |
Your haplogroup
Here is a closeup of the left of the screen:
 |
| Click to go to YFull public mtDNA tree |
Click on it to be taken to your placement in the YFull MTree.
The above screen shows a portion of the public YFull mtDNA tree. It shows the haplogroups and the individual samples within each haplogroup.
All DNA samples are identified only by an ID number.
The ones beginning with YF are from people who submitted their results. The others are from scientific studies.
You will see the notation "New" next to samples that have recently been added.
You can click on any haplogroup name to see just that portion of the mtDNA tree, or you can click on the tabs above to see earlier haplogroups.
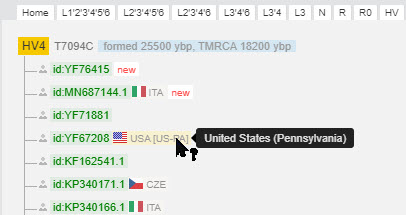
You can scan up and down the tree to any position. I did this, so in the example below we are seeing results from haplogroup HV4 because it is quite different from what we saw in haplogroup H1j2a.
 |
| Results from haplogroup HV4 |
Notice that in this case all of the scientific samples have countries of origin.
Next to the country flag you see the name of the country and maybe even a region within the country. Hover your cursor over any abbreviation to see a more full description.
In the above image you also see one ID that is shown in pink [id:MHper207]. This is an ancient sample, and you will see "age" listed next to it.
Hover your cursor over "age" to see more:
 |
| Age of ancient DNA sample |
You can see that this sample is about 950 years old (between 850 and 1050 years before present).
It is shocking to see how much the information has changed.
Here's what it looked like in 2020:
There are now many new subgroups. In the above example, the sample from Pennsylvania has recently been reassigned to haplogroup HV4e.
The MTree is the only information available to the general public.
Your mtDNA mutations
Use the MReport if you want to see a list of your mutations in one quick screen.
As mentioned above, your results were compared to two reference sequences. Both are shown on the MReport.
Here is an example of a complete list of mutations:
The first tab shows your RSRS results. The results are from the three regions of the mitochondria: the hypervariable regions that mutate more frequently (HVR1 and HVR2), and the Coding Region (CR).
The second tab shows your rCRS results.
The third tab, MTree matches, shows how each mutation was matched to other samples to determine your haplogroup.
In the example above the mutation G316A put you within haplogroup H1j2a, and mutation A15052G put you in haplogroup H1j2a1.
The mutation A302ACC is not yet on the MTree.
These haplogroups continue to evolve.
Age estimation
How old is your haplogroup? Click Age estimation.
 |
| Age estimation |
You will now see an approximate age for your nearest haplogroups. These haplogroups and age estimates change as more samples are added to the database.
 |
| YFull age estimation |
The additional tabs are + Known SNPs, + Novels, x Known SNPs, and x Novels. These are the known and novel SNPs that were used to estimate the haplogroup age, and the known and novel SNPs that were not used to calculate the age.
Mitochondrial DNA matches at YFull
Here is where you can start to find our more about the individual people who share your haplogroup.
Below is part of a list of matches.
The match list shows
- The most recent haplogroups [called the MRCA (Most Recent Common Ancestor) branch]
- An estimated time to the most recent common ancestor (TMRCA -- this is a very broad estimate)
- The most distant ancestor of the person submitting the DNA sample. This will not be shown for scientific samples.
- The country of origin if reported
- The sample ID of the person who took the test
- Private Message link
You will not receive any identifying information about your matches. For example, you will not receive an email address, but you will be able to contact anyone in your match list or in your groups, no matter how distant the match is, by using the Private Message (PM) envelope.
The PM column is listed next to the YFull ID column. You will see a link symbol
You will see an envelope
In the above list of matches, the first match is from an individual submitter. We know this because the sample begins with YF.
Unfortunately, this person did not include any information about the maternal origins. You can click the PM envelope to contact this person.
When you click the PM envelope, you will see something like this:
 |
| Send Private Message (PM) |
Again, in the above match list, the scientific samples both have countries of origin, and one of them is from an ancient sample.
An ancient sample sounds fascinating. If you click the link next to the scientific sample ID MF498722.1, it takes you to the GenBank record. Below is the first part of it.
To me, the most interesting piece of information is where to find more about the study of this sample.
The GenBank record shows the name of the article in which this sample appeared: "Female exogamy and gene pool diversification at the transition from the Final Neolithic to the Early Bronze Age in central Europe."
How do you find this article?
YFull has a list of mitochondrial DNA articles for samples found in their database. Enter the sample ID into the YFull Paper Search.
The link to the article will appear below the search box.
Click the name of the article to see an abstract and find how to access the full Article.
In the YFull Paper Search, you not only see the name of the article, but there is also a link to the samples.
Here is a partial list of the samples:
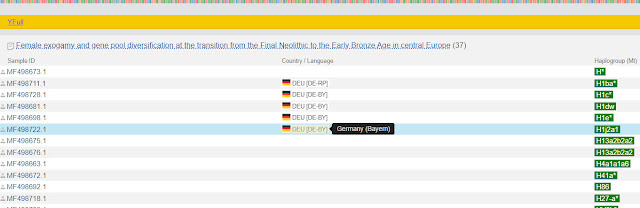 |
| List of YFull samples from scientific study |
Notice that the scientific study did not list the location of all samples, but it did for sample MF498722.1.
Although the mitochondrial DNA match list gives the country of origin as Germany, this list tells us more specific information. The sample came from the region of Bavaria.
Let's look at the Mt matches for another sample.
Here is the partial list of matches for another person:
In this case the first line is from an individual submitter. This person entered information about the most distant ancestor in the maternal line.
Two scientific samples have countries of origin, and one does not.
YFull mtDNA Groups
Your match list does not let you compare mutations between you and your matches. For that you will need to join a YFull mtDNA group.
So now we get to the best part -- YFull mtDNA groups.
Mitochondrial DNA comparisons
After you click on Groups | Mt, you will see a list of groups. After you join a group, you will be able to make great comparisons.
To see the mtDNA comparison report for your group, go to Mt-Results, and click on MReport.
Here are the first three samples from page 3 of the mtDNA group for haplogroup H1j:
Because of privacy concerns, I think it's important to address what you DON'T see on the MReport before we get to what you DO see.
First, you will not see any names. There is no identifying information of any kind, either in the match list or in the mtDNA groups. Your DNA sample will be identified only by your YFull ID number.
The Most Distant Ancestor column is not displayed in mtDNA groups. This is because, unlike in Y-DNA groups where men can have the same surname for multiple generations, the surname in a mtDNA group only represents one generation of the ancestry.
You can see the name of the most distant ancestor in your match list and on the maps [shown below] but it is not displayed in the MReport.
So, for any close match, compare the ID number in the group results with the ID number on your match list to find information about the most distant ancestor. If there is no information about the most distant maternal ancestor contact the match using the PM envelope.
Here's what you see on the MReport
The MReport has the following columns
- The YFull ID
- The PM column
- The country and region of origin
- The haplogroup
- The list of mutations that are on the YFull Mitochondrial tree in the approximate order in which the mutations occurred
- The mutations that are not yet on the tree (shown to the right in blue)
In YFull mtDNA groups the complete sequence is compared, but only the mutations relevant to the haplogroup are displayed.
Some mutations are not on the YFull tree because there aren't enough closely-matching samples in that subclade to determine the order of the mutations.
 |
| Mutations on MTree and those not yet on MTree |
The second sample above has one mutation that has not yet been placed on the YFull mtDNA tree.
Search for mutations
You can search for a mutation to see if any samples in your group have the mutation. There are two ways to do this:
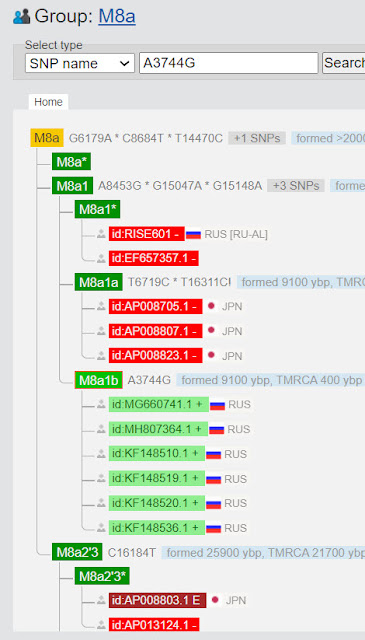
1. Search by position. Here you enter only the position, not any mutations. We are searching for position 3744.
In the example above we see that the samples within haplogroup M8a1b have the mutation G at this position, and others do not.
2. You can also search for a position or a particular mutation in the group MTree.
The group MTree shows only the samples that have been added to your group. You can compare your group MTree to the official YFull MTree to see if your group is missing any samples.
Here's how the group MTree appears before you search for a mutation:
 |
| Group MTree |
Now I will search for a mutation that everybody in haplogroup U5 should have. This mutation is not listed on the group MReport because it occurred before haplogroup U5b1c was formed.
Now let's search for a mutation that only some people will have:
In this example, only people in subclade M8a1b have the A3744G mutation.
The list of mutations on the MReports will be different in other groups
At YFull there is a U5 group as well as a U5b1c group. U5b1c is a subclade of haplogroup U5. The display of mutations on the MReport of the two groups is different.
The mutations in the U5 group include only those that occurred within haplogroup U5. More ancient mutations are not included.
Here is the list of mutations from a scientific sample in the U5 mtDNA group:
The same sample shows fewer mutations in the U5b1c group. Mutations that occurred when Haplogroup U5b1c was formed, or before, are not listed.
Here we can focus on only the mutations that are relevant to this subclade.
| List of mutations in YFull U5b1c mtDNA group |
More information about scientific samples
You can find much more information about scientific samples besides the name of the scientific article. For example, most of the ID numbers end in .1. If you see an ID number like AY519497.2, you may wonder why does it end in ".2"?
The arrow in the above image has now been changed to a link. Click the link, and you will be taken to the GenBank entry.
 |
| Scientific sample revision |
As you can see, the ".2" is a revised version of a sample that was previously named AY519497.1.
Here's another question. Sample DQ489511.1 appears in the U5 group under the subclade U5b2b3-a1b, but it is missing mutations that are shared by others within that haplogroup.
Click the link next to the ID number, and the GenBank entry reveals why this sample is missing some mutations:
 |
| Partial Genome in GenBank |
The test for this sample was only a partial genome. Perhaps someday it can be upgraded to version ".2"!
Ancestral origins from scientific samples
It is fascinating to see how the scientific samples contribute to our understanding of our ancestral origins. Here is a portion of the U5a1b1h haplogroup at YFull:
They are from individual submissions and from different scientific studies. They are all from the same region.
Here is the U5b1b1b group. They are from a completely different part of the world:
Here is the U5b1b1b group. They are from a completely different part of the world:
 |
| Countries of origin |
And here's a third subclade of haplogroup U5 that is more diverse:
 |
| Countries of origin |
As you can see in the three above images, these haplogroups are all subclades of the larger haplogroup U5. The ancestral origins continue to get more specific in the subclades.
As more scientific, and especially ancient, samples are entered into the YFull database we may be able to identify mutations with the region where they occurred.
Your results will just keep getting better.
The best feature of YFull mtDNA groups:
mtDNA maps
In addition to being able to compare mutations in mtDNA groups, you can also begin to find common ancestral origins which are displayed in the mtDNA maps.
To view the map, go to Mt-Results, then click Map. You must select a subgroup, or the map will not display results. For most groups select All.
When the map is displayed, zoom in to focus on your region of interest.
Notice that the map displays in the native characters and language of each country. The pin below is from a region near the border of Russia and China.
When you click on the pin you can see the YFull ID number and the haplogroup. This RISE601 is a scientific sample.
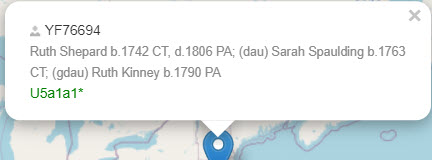
Even better, if you have entered the coordinates and the name and place of your maternal ancestor, it will display on the map.
In this case, all that was known about the ancestor was that her surname was Wang, and her place of birth was Linzi in the province of Shandong.
You can see how much better it gets as you add more information about your ancestor:
 |
| Ancestor with names, dates, and places |
The field size limits how much you can enter, but you can actually get quite creative:
Now imagine how great this will be when everybody does this! Scientific samples as well as individual submissions are included in the map.
How to start a mtDNA group at YFull
What do you do if there is no mtDNA group for your haplogroup?
Start one!
Look at the YFull mtDNA tree, and select a haplogroup that has a manageable number of scientific samples.
Start with your own haplogroup, then scroll up to a larger one if your haplogroup only has a few samples.
You could also go to the MTree to select a haplogroup:
It can be difficult to find a haplogroup by scrolling down the online MTree because some of the grouping seems completely illogical.
Look at the example below:
 |
| Subclades of haplogroup M8 |
Why are haplogroups CZ, C, and Z listed as subclades of haplogroup M8? We would expect M8a, M8b, etc. but not C and Z!
This happens because haplogroups are named when they are found. Later someone discovers that the haplogroup belongs somewhere else in the mitochondrial DNA tree.
For example, haplogroup C was named before haplogroup M8, and it was later discovered that haplogroup C was actually a subgroup of haplogroup M8. Haplogroup C retained its original name, but it is now placed under M8.
In another example, haplogroup K is a subgroup of haplogroup U8b.
For these reasons it is often easier to navigate to your haplogroup by clicking "Hg and SNPs" rather than scrolling through the MTree.
When you choose a haplogroup name for your new group, try to select the largest subclade that is reasonable for you.
You don't want to have to add 10,000 scientific samples to your group, but you also don't want a group that has only 5 samples because you won't have much to compare.
For example if your haplogroup is K2b1a1a1, you might not want to start a group for haplogroup K because it's a large group.
On the other hand, if you think that K2 or K2b would be a reasonably-sized group for you, send a message to YFull stating something like, "I would like to start a mtDNA group for haplogroup K2."
Your new group will be quickly formed.
Group administrator options
If you are not a group administrator you will see the following options in mtDNA groups:
| Group options |
If you are an administrator you will see an additional tab called GAP for the Group Administrator.
| Group administrator options |
The options in the GAP administrator tab are shown below.
When you start your group you may want to edit the group goals, group news, and group background. You don't need to add any of these, but they are places where you can add value to your group members.
I am going to edit the group news.
Here's how the group news looks now. You can copy this into your own group. Change the name of the haplogroup from HV4 to the name of your haplogroup. Then save.
 |
| Group news description at YFull |
I administer several mtDNA groups. I guess you could call me an mtDNA freak! I have used all kinds of different descriptions in the "About this group" section. Here's one I used for Group background:
Again, you can enter any description you want, or even none at all.
How to add the scientific samples
The administrator of a YFull group can add sequences from scientific samples to the group. You will be able to instantly compare your results to those of scientific samples.
Click "Add scientific sample".
 |
| Add scientific sample to group |
Open the MTree and copy the ID of scientific samples.
It is easiest if you have two screens open so that you can copy from the MTree and paste into your group.
Below I have copied and pasted, but have not yet clicked Submit. The screen shows the sample I have just pasted and the one I previously added.
Once you have added your scientific samples, you're ready to go!
You may want to go to your mtDNA match list and contact your matches using the Private Message Link. Tell them about your new group and invite them to join.
mtDNA groups are automatically sorted by haplogroup, so it requires no effort on your part. You can, however, create subgroups.
Reviewing requests to join your group
You will periodically receive an email when a new person wants to join your group. Here is an example of an email.
Click the link to see if this person is within your haplogroup. In the image below, I have removed the name and ID.
 |
| Review request to join group |
This person belongs in haplogroup U5, not in A2. I can choose to refuse any of these requests, and tell them why.
 |
| Review group Join request |
 |
| Email notice with comment from group administrator |
Most of the time you will approve the request. You do not need to add comments when you approve or deny requests, but you may want to add a comment such as "Thank you for joining!" No other efforts are necessary.
Get yourself set up at YFull:
Now that you've seen how YFull works, let's actually do it!
1. If you tested at Nebula Genomics, go to Deep Ancestry and submit your results. Your mtDNA submission is free.
Otherwise, go to yfull.com and place your order.
 |
| Order mtDNA analysis at YFull |
Fill out the information, and load your FASTA file on the next screen.
If you have already uploaded Y-DNA results for the same person from any company, you may add mtDNA results at no additional charge. Log into your YFull account, and in the menu on the left click Upload mtDNA.
Then upload your FASTA file.
2. Enter information about your most distant known maternal ancestor. Click Settings at the upper right of your Home page.
In the Account Settings tab you will find the Settings option at the right of the screen. Click Most Distant Ancestor.
You will enter your most distant-known maternal ancestor [your mother's mother's mother . . .]. Enter her name along with the approximate year and place where she was born. Be as specific as you can.
In the screenshot below, the example below the ancestor field shows the name Ann Johnson with dates of birth and death. This is not very helpful. We need a name, date, and PLACE.
Unfortunately, I do not know the county where Elizabeth Clark was born. All I know is that she was born in Pennsylvania.
Next, click Country of Origin. Enter not only the country but also the region, if known.
Set the map coordinates so that your sample will display in the precise location on the mtDNA maps.
3. Join a mtDNA group if one exists for your haplogroup.
In the menu on the left, scroll down to Groups | Mt.
 |
| YFull mtDNA Groups |
If there is no mtDNA group for your haplogroup, you can wait for someone to start one (new groups are currently being formed), or you can easily start one yourself.
4. You can learn about other relatives by finding people who are descended from any of your maternal lines. Encourage these people to test their mtDNA and to join YFull.
For example, you could encourage a cousin to take a test from Nebula Genomics. Tell her about all of the health benefits and courage her to upload her mtDNA results to YFull.
Summary
When you submit your mtDNA results to YFull, you will understand where you fit into the great tree of women.
You will make matches with other people and be able to compare mtDNA and ancestry.
You will also find your final mutations to see when you reach the end of the mtDNA trail.
Remember that your matches and haplogroups will continue to evolve.
Get started today!
What's next?
To find out more about mtDNA matches see How to make matches with Mitochondrial DNA (mtDNA)
You will see other places where you can transfer your mtDNA.